[English] 日本語
 Yorodumi
Yorodumi- EMDB-11736: Cryo-EM structure of human RNA Polymerase III elongation complex 1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11736 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | |||||||||
 Map data Map data | Elongating Human Pol III complex Map B, unsharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HUMAN / HUMAN /  TRANSCRIPTION / SHORT RNAs TRANSCRIPTION / SHORT RNAs | |||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA transcription by RNA polymerase III / RNA Polymerase III Chain Elongation / RNA Polymerase III Transcription Termination /  calcitonin gene-related peptide receptor activity / DNA/RNA hybrid binding / regulation of transcription by RNA polymerase I / RPAP3/R2TP/prefoldin-like complex / regulation of transcription by RNA polymerase III / calcitonin gene-related peptide receptor activity / DNA/RNA hybrid binding / regulation of transcription by RNA polymerase I / RPAP3/R2TP/prefoldin-like complex / regulation of transcription by RNA polymerase III /  DNA polymerase III complex / Cytosolic sensors of pathogen-associated DNA ...snRNA transcription by RNA polymerase III / RNA Polymerase III Chain Elongation / RNA Polymerase III Transcription Termination / DNA polymerase III complex / Cytosolic sensors of pathogen-associated DNA ...snRNA transcription by RNA polymerase III / RNA Polymerase III Chain Elongation / RNA Polymerase III Transcription Termination /  calcitonin gene-related peptide receptor activity / DNA/RNA hybrid binding / regulation of transcription by RNA polymerase I / RPAP3/R2TP/prefoldin-like complex / regulation of transcription by RNA polymerase III / calcitonin gene-related peptide receptor activity / DNA/RNA hybrid binding / regulation of transcription by RNA polymerase I / RPAP3/R2TP/prefoldin-like complex / regulation of transcription by RNA polymerase III /  DNA polymerase III complex / Cytosolic sensors of pathogen-associated DNA / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / RNA Polymerase III Abortive And Retractive Initiation / DNA polymerase III complex / Cytosolic sensors of pathogen-associated DNA / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / RNA Polymerase III Abortive And Retractive Initiation /  RNA polymerase III activity / positive regulation of innate immune response / Abortive elongation of HIV-1 transcript in the absence of Tat / nucleobase-containing compound metabolic process / MicroRNA (miRNA) biogenesis / RNA Polymerase I Transcription Termination / FGFR2 alternative splicing / Viral Messenger RNA Synthesis / Signaling by FGFR2 IIIa TM / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / termination of RNA polymerase III transcription / PIWI-interacting RNA (piRNA) biogenesis / RNA polymerase III activity / positive regulation of innate immune response / Abortive elongation of HIV-1 transcript in the absence of Tat / nucleobase-containing compound metabolic process / MicroRNA (miRNA) biogenesis / RNA Polymerase I Transcription Termination / FGFR2 alternative splicing / Viral Messenger RNA Synthesis / Signaling by FGFR2 IIIa TM / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / termination of RNA polymerase III transcription / PIWI-interacting RNA (piRNA) biogenesis /  RNA polymerase I activity / mRNA Splicing - Minor Pathway / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA polymerase I activity / mRNA Splicing - Minor Pathway / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance /  transcription initiation at RNA polymerase III promoter / tRNA transcription by RNA polymerase III / RNA Polymerase I Transcription Initiation / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / RNA polymerase II transcribes snRNA genes / Processing of Capped Intron-Containing Pre-mRNA / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / neuropeptide signaling pathway / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / transcription initiation at RNA polymerase III promoter / tRNA transcription by RNA polymerase III / RNA Polymerase I Transcription Initiation / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / RNA polymerase II transcribes snRNA genes / Processing of Capped Intron-Containing Pre-mRNA / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / neuropeptide signaling pathway / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat /  RNA polymerase I complex / transcription by RNA polymerase I / RNA polymerase I complex / transcription by RNA polymerase I /  RNA polymerase III complex / transcription by RNA polymerase III / Formation of HIV elongation complex in the absence of HIV Tat / RNA polymerase III complex / transcription by RNA polymerase III / Formation of HIV elongation complex in the absence of HIV Tat /  RNA polymerase II, core complex / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / Inhibition of DNA recombination at telomere / positive regulation of interferon-beta production / mRNA Splicing - Major Pathway / acrosomal vesicle / RNA polymerase II, core complex / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / Inhibition of DNA recombination at telomere / positive regulation of interferon-beta production / mRNA Splicing - Major Pathway / acrosomal vesicle /  protein-DNA complex / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase I Promoter Escape / Transcriptional regulation by small RNAs / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / protein-DNA complex / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase I Promoter Escape / Transcriptional regulation by small RNAs / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / Transcription-Coupled Nucleotide Excision Repair (TC-NER) /  ribonucleoside binding / Formation of TC-NER Pre-Incision Complex / ribonucleoside binding / Formation of TC-NER Pre-Incision Complex /  fibrillar center / DNA-directed 5'-3' RNA polymerase activity / fibrillar center / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / DNA-directed RNA polymerase / Activation of anterior HOX genes in hindbrain development during early embryogenesis / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER /  single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding /  double-stranded DNA binding / defense response to virus / Estrogen-dependent gene expression / cell population proliferation / transcription by RNA polymerase II / double-stranded DNA binding / defense response to virus / Estrogen-dependent gene expression / cell population proliferation / transcription by RNA polymerase II /  nucleic acid binding / protein stabilization / nucleic acid binding / protein stabilization /  nuclear body / nuclear body /  protein dimerization activity / protein dimerization activity /  nucleotide binding / intracellular membrane-bounded organelle / nucleotide binding / intracellular membrane-bounded organelle /  innate immune response / innate immune response /  centrosome / DNA-templated transcription / centrosome / DNA-templated transcription /  chromatin binding / magnesium ion binding / chromatin binding / magnesium ion binding /  DNA binding / zinc ion binding / DNA binding / zinc ion binding /  nucleoplasm nucleoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
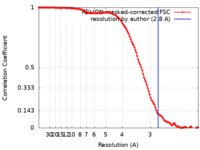
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Girbig M / Misiaszek AD | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Authors: Mathias Girbig / Agata D Misiaszek / Matthias K Vorländer / Aleix Lafita / Helga Grötsch / Florence Baudin / Alex Bateman / Christoph W Müller /   Abstract: RNA polymerase III (Pol III) synthesizes transfer RNAs and other short, essential RNAs. Human Pol III misregulation is linked to tumor transformation, neurodegenerative and developmental disorders, ...RNA polymerase III (Pol III) synthesizes transfer RNAs and other short, essential RNAs. Human Pol III misregulation is linked to tumor transformation, neurodegenerative and developmental disorders, and increased sensitivity to viral infections. Here, we present cryo-electron microscopy structures at 2.8 to 3.3 Å resolution of transcribing and unbound human Pol III. We observe insertion of the TFIIS-like subunit RPC10 into the polymerase funnel, providing insights into how RPC10 triggers transcription termination. Our structures resolve elements absent from Saccharomyces cerevisiae Pol III such as the winged-helix domains of RPC5 and an iron-sulfur cluster, which tethers the heterotrimer subcomplex to the core. The cancer-associated RPC7α isoform binds the polymerase clamp, potentially interfering with Pol III inhibition by tumor suppressor MAF1, which may explain why overexpressed RPC7α enhances tumor transformation. Finally, the human Pol III structure allows mapping of disease-related mutations and may contribute to the development of inhibitors that selectively target Pol III for therapeutic interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11736.map.gz emd_11736.map.gz | 113.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11736-v30.xml emd-11736-v30.xml emd-11736.xml emd-11736.xml | 54.5 KB 54.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11736_fsc.xml emd_11736_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11736.png emd_11736.png | 74 KB | ||
| Masks |  emd_11736_msk_1.map emd_11736_msk_1.map | 144.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11736.cif.gz emd-11736.cif.gz | 11.4 KB | ||
| Others |  emd_11736_additional_1.map.gz emd_11736_additional_1.map.gz emd_11736_additional_2.map.gz emd_11736_additional_2.map.gz emd_11736_additional_3.map.gz emd_11736_additional_3.map.gz emd_11736_additional_4.map.gz emd_11736_additional_4.map.gz emd_11736_additional_5.map.gz emd_11736_additional_5.map.gz emd_11736_half_map_1.map.gz emd_11736_half_map_1.map.gz emd_11736_half_map_2.map.gz emd_11736_half_map_2.map.gz | 112.9 MB 118.8 MB 98.2 MB 10.5 MB 5.5 MB 113.8 MB 113.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11736 http://ftp.pdbj.org/pub/emdb/structures/EMD-11736 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11736 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11736 | HTTPS FTP |
-Related structure data
| Related structure data |  7ae1MC  7a6hC  7ae3C  7aeaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10697 (Title: Cryo-EM structures of human RNA Polymerase III / Data size: 3.3 TB EMPIAR-10697 (Title: Cryo-EM structures of human RNA Polymerase III / Data size: 3.3 TBData #1: LZW-TIFF compressed multiframe micrographs of human RNA Pol III EC [micrographs - multiframe] Data #2: Initially picked particles of human RNA Pol III EC [picked particles - single frame - processed] Data #3: Polished particles of human RNA Pol III EC (final reconstruction) [picked particles - single frame - processed] Data #4: LZW-TIFF compressed multiframe micrographs of human RNA Pol III apo [micrographs - multiframe] Data #5: Initially picked particles of human RNA Pol III apo [picked particles - single frame - processed] Data #6: Polished particles of human RNA Pol III apo (final reconstruction) [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11736.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11736.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Elongating Human Pol III complex Map B, unsharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11736_msk_1.map emd_11736_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
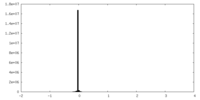
| Projections & Slices |
| ||||||||||||
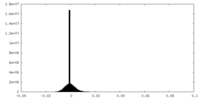
| Density Histograms |
-Additional map: Elongating Human Pol III complex Map B, locally...
| File | emd_11736_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Elongating Human Pol III complex Map B, locally sharpened with LocalDeblur | ||||||||||||
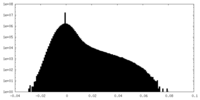
| Projections & Slices |
| ||||||||||||
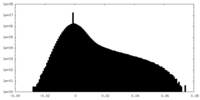
| Density Histograms |
-Additional map: Elongating Human Pol III complex Map B1, from...
| File | emd_11736_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Elongating Human Pol III complex Map B1, from RELION multi-body refinement, body_001 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Elongating Human Pol III complex Map B2, from...
| File | emd_11736_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Elongating Human Pol III complex Map B2, from RELION multi-body refinement, body_002 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Elongating Human Pol III complex Map B1, from...
| File | emd_11736_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Elongating Human Pol III complex Map B1, from RELION multi-body refinement, body_001, sharpened with Density Modification | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Elongating Human Pol III complex Map B2, from...
| File | emd_11736_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Elongating Human Pol III complex Map B2, from RELION multi-body refinement, body_002, sharpened with Density Modification | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_11736_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_11736_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Human RNA Polymerase III Elongation Complex 1
+Supramolecule #1: Human RNA Polymerase III Elongation Complex 1
+Supramolecule #2: Human RNA Polymerase
+Supramolecule #3: RNA, DNA
+Macromolecule #1: DNA-directed RNA polymerase III subunit RPC1
+Macromolecule #2: DNA-directed RNA polymerase III subunit RPC2
+Macromolecule #3: DNA-directed RNA polymerases I and III subunit RPAC1
+Macromolecule #4: DNA-directed RNA polymerase III subunit RPC9
+Macromolecule #5: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #6: DNA-directed RNA polymerases I, II, and III subunit RPABC2
+Macromolecule #7: DNA-directed RNA polymerase III subunit RPC8
+Macromolecule #8: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #9: DNA-directed RNA polymerase III subunit RPC10
+Macromolecule #10: DNA-directed RNA polymerases I, II, and III subunit RPABC5
+Macromolecule #11: DNA-directed RNA polymerases I and III subunit RPAC2
+Macromolecule #12: DNA-directed RNA polymerases I, II, and III subunit RPABC4
+Macromolecule #13: DNA-directed RNA polymerase III subunit RPC5
+Macromolecule #14: DNA-directed RNA polymerase III subunit RPC4
+Macromolecule #15: DNA-directed RNA polymerase III subunit RPC3
+Macromolecule #16: DNA-directed RNA polymerase III subunit RPC6
+Macromolecule #17: DNA-directed RNA polymerase III subunit RPC7
+Macromolecule #18: RNA
+Macromolecule #19: Non-template DNA
+Macromolecule #20: Template-DNA
+Macromolecule #21: ZINC ION
+Macromolecule #22: IRON/SULFUR CLUSTER
+Macromolecule #23: MAGNESIUM ION
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Wait time 10 s Blot force 4 Blot time 4 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.4 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z
Z Y
Y X
X