[English] 日本語
 Yorodumi
Yorodumi- EMDB-1134: Polymorphism and double hexamer structure in the archaeal minichr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1134 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
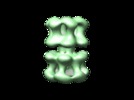
| Title | Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. | |||||||||
 Map data Map data | test map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | MCM domain / DNA replication initiation Function and homology information Function and homology information | |||||||||
| Biological species |    Methanothermobacter thermautotrophicus (archaea) Methanothermobacter thermautotrophicus (archaea) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / cryo EM /  negative staining / Resolution: 25.0 Å negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Gomez-Llorente Y / Fletcher RJ / Chen XS / Carazo JM / San Martin C | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2005 Journal: J Biol Chem / Year: 2005Title: Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. Authors: Yacob Gómez-Llorente / Ryan J Fletcher / Xiaojiang S Chen / José M Carazo / Carmen San Martín /  Abstract: Methanobacterium thermoautotrophicum minichromosome maintenance complex (mtMCM), a cellular replicative helicase, is a useful model for the more complex eukaryotic MCMs. Biochemical and ...Methanobacterium thermoautotrophicum minichromosome maintenance complex (mtMCM), a cellular replicative helicase, is a useful model for the more complex eukaryotic MCMs. Biochemical and crystallographic evidence indicates that mtMCM assembles as a double hexamer (dHex), but previous electron microscopy studies reported only the presence of single heptamers or single hexamers (Pape, T., Meka, H., Chen, S., Vicentini, G., Van Heel, M., and Onesti, S. (2003) EMBO Rep. 4, 1079-1083; Yu, X., VanLoock, M. S., Poplawski, A., Kelman, Z., Xiang, T., Tye, B. K., and Egelman, E. H. (2002) EMBO Rep. 3, 792-797). Here we present the first three-dimensional electron microscopy reconstruction of the full-length mtMCM dHex in which two hexamers contact each other via the structurally well defined N-terminal domains. The dHex has obvious side openings that resemble the side channels of LTag (large T antigen). 6-fold and 7-fold rings were observed in the same mtMCM preparation, but we determined that assembly as a double ring favors 6-fold structures. Additionally, open rings were also detected, which suggests a direct mtMCM loading mechanism onto DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1134.map.gz emd_1134.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1134-v30.xml emd-1134-v30.xml emd-1134.xml emd-1134.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  1134.gif 1134.gif | 10.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1134 http://ftp.pdbj.org/pub/emdb/structures/EMD-1134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1134 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1134.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1134.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | test map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Archaeal helicase MCM from Methanobacterium thermoautotrophicum
| Entire | Name: Archaeal helicase MCM from Methanobacterium thermoautotrophicum |
|---|---|
| Components |
|
-Supramolecule #1000: Archaeal helicase MCM from Methanobacterium thermoautotrophicum
| Supramolecule | Name: Archaeal helicase MCM from Methanobacterium thermoautotrophicum type: sample / ID: 1000 / Oligomeric state: homododecamer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 900 KDa / Theoretical: 900 KDa / Method: gel filtration |
-Macromolecule #1: minichromosome maintenance protein
| Macromolecule | Name: minichromosome maintenance protein / type: protein_or_peptide / ID: 1 / Name.synonym: MCM / Number of copies: 12 / Oligomeric state: dodecamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Methanothermobacter thermautotrophicus (archaea) / Strain: Delta H / synonym: Methanobacterium thermoautotrophicum Methanothermobacter thermautotrophicus (archaea) / Strain: Delta H / synonym: Methanobacterium thermoautotrophicum |
| Molecular weight | Experimental: 75.6 MDa / Theoretical: 75.6 MDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | GO: DNA replication initiation / InterPro: MCM domain |
-Experimental details
-Structure determination
| Method |  negative staining, negative staining,  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50 mM Tris.HCl pH 8.0, 1 mM DTT, 50 mM to 1M NaCl |
| Staining | Type: NEGATIVE / Details: 2% uranyl acetate |
| Grid | Details: glow discharged, collodion/carbon coated copper grids |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1200EXII |
|---|---|
| Electron beam | Acceleration voltage: 80 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 5.6 mm / Nominal magnification: 60000 Bright-field microscopy / Cs: 5.6 mm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: side entry / Specimen holder model: OTHER |
| Alignment procedure | Legacy - Astigmatism: visually corrected at 100,000x |
| Details | he make and model of the microscope. Jeol 1200 EX-II. Standard Jeol 1200 holder |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
- Image processing
Image processing
| Final angle assignment | Details: theta between 75 and 90, phi between 0 and 360 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: SPIDER, XMIPP / Details: reconstructed with ART / Number images used: 1200 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: SPIDER, XMIPP / Details: reconstructed with ART / Number images used: 1200 |
-Atomic model buiding 1
| Software | Name: Amira |
|---|
 Movie
Movie Controller
Controller