-Search & filter documents
 FAQ,
FAQ,  News,
News,  Information,
Information,  EM Navigator,
EM Navigator,  Yorodumi,
Yorodumi,  Omokage search,
Omokage search,  Services,
Services,  All docs
All docs
-News (9)
+Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
+Jun 16, 2017. Omokage search with filter
Omokage search with filter
Result of Omokage search can be filtered by keywords and the database types
Related info.: Omokage search
Omokage search
+Apr 13, 2016. Omokage search got faster
Omokage search got faster
- The computation time became ~1/2 compared to the previous version by re-optimization of data accession.
- Enjoy "shape similarity" of biomolecules, more!
Related info.: Omokage search
Omokage search
+Mar 3, 2016. Presentation PDF file for IPR seminar on Feb 19.
Presentation PDF file for IPR seminar on Feb 19.
External links: IPR seminar Feb 19th, 2016 /
IPR seminar Feb 19th, 2016 /  Presentation PDF
Presentation PDF
+Dec 4, 2015. The article about Omokage search is published online
The article about Omokage search is published online
Omokage search: shape similarity search service for biomolecular structures in both the PDB and EMDB. Suzuki Hirofumi, Kawabata Takeshi, and Nakamura Haruki, Bioinformatics. (2015) btv614
Related info.: Omokage search /
Omokage search /  gmfit
gmfit
External links: Main text (HTML, Open Access) /
Main text (HTML, Open Access) /  Supplementray data (PDF, Open Access)
Supplementray data (PDF, Open Access)
+Nov 28, 2015. Omokage search starts to support SASBDB models
Omokage search starts to support SASBDB models
- Models data in SASBDB are included in the Omokage search database.
- SASBDB is a databank for small angle scattering data.
- A SASBDB model can be used as a search query.
- The search result may include SASBDB models.
Related info.: Omokage search /
Omokage search /  SASBDB /
SASBDB /  Comparison of 3 databanks
Comparison of 3 databanks
+Sep 22, 2014. New service: Omokage search
New service: Omokage search
- Shape similarity search service, Omokage search has started.
- 形状類似検索「Omokage検索」を開始しました。
Related info.: Omokage search
Omokage search
-FAQ (3)
+Q: How do you make the images for the structure data? What do their colors mean?
A: Images of EMDB entries are made for EM Navigator, and ones of PDB entries are for Yorodumi. There are several patterns of coloring.
See following items for the details.
Related info.: Q: How do you make the movies in EM Navigator? /
Q: How do you make the movies in EM Navigator? /  Q: How do you make the images of EMDB entries in Yorodumi, Omokage, etc.? /
Q: How do you make the images of EMDB entries in Yorodumi, Omokage, etc.? /  Q: How do you make the images of PDB entries in Yorodumi, Omokage, etc.?
Q: How do you make the images of PDB entries in Yorodumi, Omokage, etc.?
+Q: How do you make the images of EMDB entries in Yorodumi, Omokage, etc.?
A: in semi-automatic process with UCSF Chimera
- Method:
- We make been creating the images and movies for structure entries in EM Navigator using UCSF Chimera in semi-automatic process.
- Orientation:
- The structures are manually rotated to be seen in "good" orientation (usually, figures of the paper is refererd), or in similar orientation with similar structure data.
- For the case of icosahedral symmetry structure data, they are viewed normal to their five-fold axis, while typical view of such the structure data are along the two-fold or three-fold axes. This is to unify the orientation with structure data with pseudo-icosahedral symmetry, such as bacteriophage with tail structures.
- Color:
- In EM Navigator, multiple images are made for a single entry. In Omokage search, image of "colored surface view" are shown.
- The surface are colored by height, distance, or cylindrical radius. See the detail page for the coloring of particular images.
Related info.: Q: How do you make the images of PDB entries in Yorodumi, Omokage, etc.? /
Q: How do you make the images of PDB entries in Yorodumi, Omokage, etc.? /  Q: How do you make the movies in EM Navigator? /
Q: How do you make the movies in EM Navigator? /  Q: How do you make the images for the structure data? What do their colors mean?
Q: How do you make the images for the structure data? What do their colors mean?
External links: UCSF Chimera Home Page
UCSF Chimera Home Page
+Q: How do you make the images of PDB entries in Yorodumi, Omokage, etc.?
A: We are making them by full-automatic process using Jmol. Their styles are depend on the data type
- Orientation:
- Icosahedral assembly: Original orientation.
- Helical assembly: 6 orientation (+/- of X, Y, Z direction) images are generated in jpeg format. Then, the largest file is chosen. (large JPEG => many dots/colors)
- Others: by "rotate best" command of Jmol.
- Ribosomes: by "rotate best" command of Jmol. Only RNA coordinates are used for the orientation.
- Color:
- monomer AUs: by N->C (blue->red) rainbow
- BUs of monomer AUs: by "color molecule" (Jmol's coloring)
- multimer AUs and thier BUs (including monomer BUs): by "color chain". For this coloring, Jmol uses not a rainbow color but the colors determined by the chain-ID.
- Problems:
- BU models are generated by Jmol's system. It works well for the most data, but there are some exception.
- Checking and fixing processes of the many (>200,000) images are in progress. There are still some wrong images.
Related info.: Q: How do you make the images of EMDB entries in Yorodumi, Omokage, etc.?
Q: How do you make the images of EMDB entries in Yorodumi, Omokage, etc.?
External links: Jmol: an open-source Java viewer for chemical structures in 3D
Jmol: an open-source Java viewer for chemical structures in 3D
-Information (9)
+Omokage search
Search structure by SHAPE

URL: https://pdbj.org/omokage/
- "Omokage search" is a shape similarity search service for 3D structures of macromolecules. By comparing global shapes, and ignoring details, similar-shaped structures are searched.
- The search is performed ageinst >200,000 structure data, which consists of EMDB map data, PDB coordinates (deposited units (asymmetric units, usually), PDB biological units, and SASBDB mdoels).
- For the search query, you can use either a data in the PDB/EMDB/SASBDB or your original model.
- Supported formats are PDB (atomic model, SAXS bead model, etc.) and CCP4/MRC map (3D density map).
- Shape comparison is performed by iDR profile method, which uses 1D-distance profiles of super-simplified models generated by vector quantizaion in Situs package.
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Sep 22, 2014. New service: Omokage search /
Sep 22, 2014. New service: Omokage search /  gmfit /
gmfit /  Re-ranking by gmfit
Re-ranking by gmfit
+Yorodumi Docs
+Covid-19 info
Page for Covid-19 featured contents

URL: https://pdbj.org/emnavi/covid19.php
Related info.: Comparison of 3 databanks /
Comparison of 3 databanks /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
+F&H Search
Function & Homology similarity search
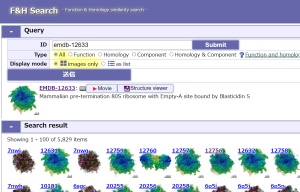
URL: https://pdbj.org/emnavi/fh-search.php
Based on the similarity of Function & Homolog items related to the structure data, similar data are searched from structure databases.
Related info.: Function and homology information
Function and homology information
+gmfit
A program for fitting subunits into density map of complex using GMM (Gaussian Mixture Model)

- Developed by Takeshi Kawabata in IPR, Osaka-univ.
- Used from Omokage search.
Related info.: Omokage search /
Omokage search /  Dec 4, 2015. The article about Omokage search is published online /
Dec 4, 2015. The article about Omokage search is published online /  Re-ranking by gmfit
Re-ranking by gmfit
External links: Pairwise gmfit
Pairwise gmfit
+Re-ranking by gmfit
The similarity ranking by Omokage search can be re-ordered according to correlation coefficient by gmfit, expecting incorporation of the (potential) benefits of two methods with following properties.
| Name | Method | Speed | Potential accuracy |
|---|---|---|---|
| Omokage search | iDR profile comparison | ++ sub-msec / 1 comparison | - 1D profile comparison |
| gmfit | Gaussian mixture model fitting | + sub-sec / 1 comparison | + comparison in 3D space |
Related info.: gmfit /
gmfit /  Omokage search
Omokage search
+Changes in new EM Navigator and Yorodumi
Changes in new versions of EM Navigator and Yorodumi. (Sep. 2016)
- Changes:
- New user interface unified in EM Navigator, Yorodumi & Omokage search, supporting mobile devices as well as PCs.
- New Yorodumi replace the individual data page ("Detail page" of EM Navigator in the legacy system). It is unified browser for EMDB, PDB, and SASBDB entries integrated with EM Navigator and Omokage search.
- The viewers (structure/movie viewers) appear in pop-up windows. On a PC, multiple viewer windows can be opened. On mobile devices, the viewers support touch operation.
- New features:
- Molmil, molecular structure viewer, and SurfView, surface model viewer for EMDB map data, are available to view the 3D structures. Both viewers support mobile device use.
- Yorodumi support SASBDB entries.
- Some new pages such as Yorodumi Papers, citation database of structure data entries and Yorodumi Search, cross search by keywords
Related info.: EM Navigator /
EM Navigator /  Yorodumi /
Yorodumi /  SurfView /
SurfView /  Molmil /
Molmil /  Movie viewer /
Movie viewer /  EMN Papers /
EMN Papers /  Yorodumi Papers /
Yorodumi Papers /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi
Aug 31, 2016. New EM Navigator & Yorodumi
+Information from PDBMLplus
PDBMLplus is the XML format file including additional information relating to individual proteins. Currently, PDB files are lacking a detailed description of function, experimental conditions, and the like. And then such information was included to extended XML database, named PDBMLplus.
Related info.: PDBj /
PDBj /  Function and homology information
Function and homology information
External links: PDBMLplus - PDBj helip /
PDBMLplus - PDBj helip /  About Functional Details page - PDBJ help
About Functional Details page - PDBJ help
+Yorodumi annotation
Annotation by Yorodumi/EM Navigator manager
Related info.: Q: Who make these documents? Who develop EM Navigator, Yorodumi, Omokage search, etc?
Q: Who make these documents? Who develop EM Navigator, Yorodumi, Omokage search, etc?
 Movie
Movie Controller
Controller Structure viewers
Structure viewers