2JPT
 
 | |
2V1V
 
 | |
6RPV
 
 | | Extremely stable monomeric variant of human cystatin C with single amino acid substitution | | Descriptor: | Cystatin-C | | Authors: | Zhukov, I, Rodziewicz-Motowidlo, S, Maszota-Zieleniak, M, Jurczak, P, Kozak, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
2RQS
 
 | | 3D structure of Pin from the psychrophilic archeon Cenarcheaum symbiosum (CsPin) | | Descriptor: | Parvulin-like peptidyl-prolyl isomerase | | Authors: | Zhukov, I, Jaremko, L, Jaremko, M, Mueller, J.W, Bayer, P. | | Deposit date: | 2009-11-17 | | Release date: | 2010-11-24 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the First Archaeal Parvulin Reveal a New Functionally Important Loop in Parvulin-type Prolyl Isomerases
J.Biol.Chem., 286, 2011
|
|
8ONG
 
 | |
2V31
 
 | | Structure of First Catalytic Cysteine Half-domain of mouse ubiquitin- activating enzyme | | Descriptor: | UBIQUITIN-ACTIVATING ENZYME E1 X | | Authors: | Jaremko, L, Jaremko, M, Wojciechowski, W, Filipek, R, Szczepanowski, R.H, Bochtler, M, Zhukov, I. | | Deposit date: | 2007-06-11 | | Release date: | 2008-06-24 | | Last modified: | 2018-10-24 | | Method: | SOLUTION NMR | | Cite: | Structure of First Catalytic Cysteine Half-Domain of Mouse Ubiquitin-Activating Enzyme
To be Published
|
|
2LFT
 
 | | Human prion protein with E219K protective polymorphism | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giacin, G, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-07-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism.
Biochem.J., 446, 2012
|
|
2NAR
 
 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2018-06-06 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
2LEJ
 
 | | human prion protein mutant HuPrP(90-231, M129, V210I) | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giachin, G, Raspadori, A, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Toward the Molecular Basis of Inherited Prion Diseases: NMR Structure of the Human Prion Protein with V210I Mutation.
J.Mol.Biol., 412, 2011
|
|
2LSB
 
 | | Solution-state NMR structure of the human prion protein | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giancin, G, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2012-04-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the protective effect of the human prion protein carrying the dominant-negative E219K polymorphism.
Biochem.J., 446, 2012
|
|
2LLS
 
 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
2LLT
 
 | | Post-translational S-nitrosylation is an endogenous factor fine-tuning human S100A1 protein properties | | Descriptor: | Protein S100-A1 | | Authors: | Lenarcic Zivkovic, M, Zareba-Koziol, M, Zhukova, L, Poznanski, J, Zhukov, I, Wyslouch-Cieszynska, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-09-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
J.Biol.Chem., 287, 2012
|
|
2M5X
 
 | | Novel method of protein purification for structural research. Example of ultra high resolution structure of SPI-2 inhibitor by X-ray and NMR spectroscopy. | | Descriptor: | Silk protease inhibitor 2 | | Authors: | Lenarcic Zivkovic, M, Dvornyk, A, Kludkiewicz, B, Kopera, E, Zagorski-Ostoja, W, Grzelak, K, Zhukov, I, Bal, W. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
2JT9
 
 | | NMR structure of immunosuppressory peptide containing cyclolinopeptide X and antennapedia(43-58) sequences | | Descriptor: | 5-mer immunosuppressory peptide from cyclolinopeptide X, 6-AMINOHEXANOIC ACID, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Jaremko, L, Jaremko, M, Zhukov, I, Cebrat, M. | | Deposit date: | 2007-07-21 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of immunosuppressory peptide containing cyclolinopeptide X
and antennapedia(43-58) sequences
To be Published
|
|
2JTA
 
 | |
2KUN
 
 | | Three dimensional structure of HuPrP(90-231 M129 Q212P) | | Descriptor: | Major prion protein | | Authors: | Ilc, G, Giachin, G, Jaremko, M, Jaremko, L, Zhukov, I, Plavec, J, Legname, G, Benetti, F. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human prion protein with the pathological Q212P mutation reveals unique structural features.
Plos One, 5, 2010
|
|
2L0P
 
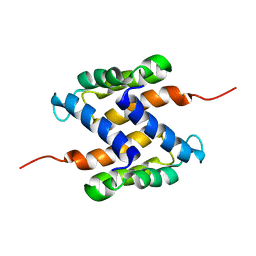 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2017-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
2LHL
 
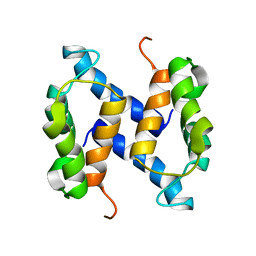 | | Chemical Shift Assignments and solution structure of human apo-S100A1 E32Q mutant | | Descriptor: | Protein S100-A1 | | Authors: | Ruszczynska-Bartnik, K, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N NMR sequence-specific resonance assignments and relaxation parameters for human apo-S100A1 E32Q mutant
To be Published
|
|
2LLU
 
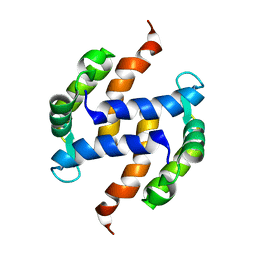 | | Post-translational S-nitrosylation is an endogenous factor fine-tuning human S100A1 protein properties | | Descriptor: | Protein S100-A1 | | Authors: | Lenarcic Zivkovic, M, Zareba-Koziol, M, Zhukova, L, Poznanski, J, Zhukov, I, Wyslouch-Cieszynska, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-09-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
J.Biol.Chem., 287, 2012
|
|
6QBJ
 
 | |
4HGU
 
 | | Crystal Structure of Galleria mellonella Silk Protease Inhibitor 2 | | Descriptor: | SODIUM ION, Silk protease inhibitor 2 | | Authors: | Krzywda, S, Jaskolski, M, Dvornyk, A, Kludkiewicz, B, Grzelak, K, Zagorski, W, Bal, W, Kopera, E. | | Deposit date: | 2012-10-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
6ROA
 
 | | Crystal structure of V57G mutant of human cystatin C | | Descriptor: | Cystatin-C | | Authors: | Orlikowska, M, Behrendt, I, Borek, D, Otwinowski, Z, Skowron, P, Szymanska, A. | | Deposit date: | 2019-05-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
6QAN
 
 | |
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
1LU0
 
 | | Atomic Resolution Structure of Squash Trypsin Inhibitor: Unexpected Metal Coordination | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Thaimattam, R, Tykarska, E, Bierzynski, A, Sheldrick, G.M, Jaskolski, M. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution structure of squash trypsin inhibitor: unexpected metal coordination.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
