4EAI
 
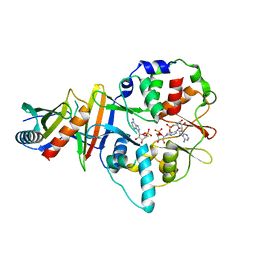 | | Co-crystal structure of an AMPK core with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAK
 
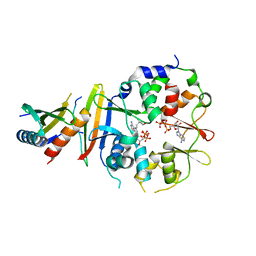 | | Co-crystal structure of an AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAJ
 
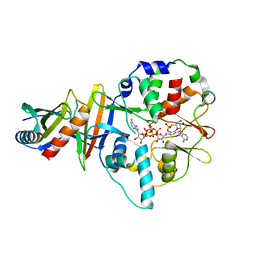 | | Co-crystal of AMPK core with AMP soaked with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAG
 
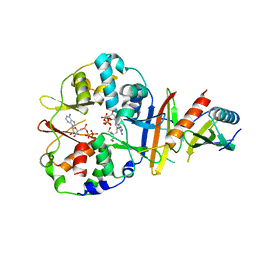 | | Co-crystal structure of an chimeric AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAL
 
 | | Co-crystal of AMPK core with ATP soaked with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4XU5
 
 | | Crystal structure of MvINS bound to a bromine-derived 14C Diacylglycerol (DAG) at 2.1A resolution | | Descriptor: | (2S)-1-[(13-bromotridecanoyl)oxy]-3-hydroxypropan-2-yl tetradecanoate, DECANE, Uncharacterized protein, ... | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU4
 
 | | Crystal structure of a mycobacterial Insig homolog MvINS from Mycobacterium vanbaalenii at 1.9A resolution | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Uncharacterized protein, nonyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU6
 
 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
8EJX
 
 | |
3RT0
 
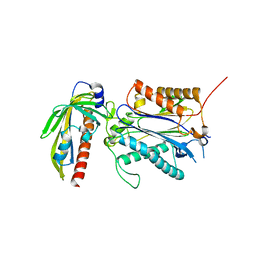 | | Crystal structure of PYL10-HAB1 complex in the absence of abscisic acid (ABA) | | Descriptor: | Abscisic acid receptor PYL10, MAGNESIUM ION, Protein phosphatase 2C 16 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
3RT2
 
 | | Crystal structure of apo-PYL10 | | Descriptor: | Abscisic acid receptor PYL10 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
4F2L
 
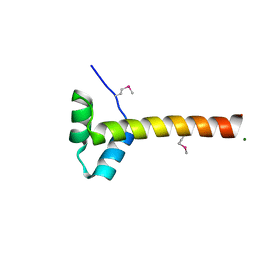 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6VXG
 
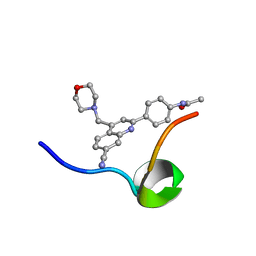 | | Structure of the C-terminal Domain of RAGE and Its Inhibitor | | Descriptor: | Advanced glycosylation end product-specific receptor, N-(4-{7-cyano-4-[(morpholin-4-yl)methyl]quinolin-2-yl}phenyl)acetamide | | Authors: | Ramirez, L, Shekhtman, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Small-molecule antagonism of the interaction of the RAGE cytoplasmic domain with DIAPH1 reduces diabetic complications in mice.
Sci Transl Med, 13, 2021
|
|
