4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
1WVR
 
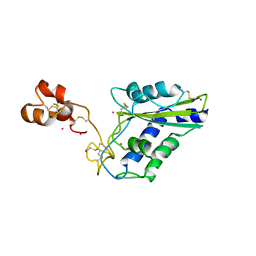 | | Crystal Structure of a CRISP family Ca-channel blocker derived from snake venom | | Descriptor: | CADMIUM ION, Triflin | | Authors: | Shikamoto, Y, Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-12-24 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a CRISP family Ca2+ -channel blocker derived from snake venom.
J.Mol.Biol., 350, 2005
|
|
2D01
 
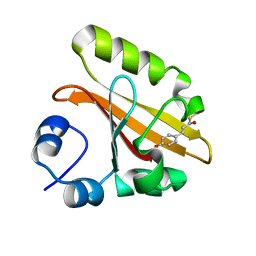 | | Wild Type Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
2D02
 
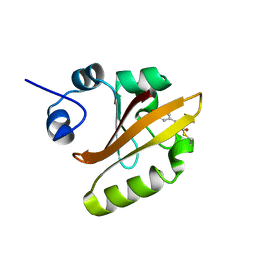 | | R52Q Mutant of Photoactive Yellow Protein, P65 Form | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Shimizu, N, Kamikubo, H, Yamazaki, Y, Imamoto, Y, Kataoka, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Crystal Structure of the R52Q Mutant Demonstrates a Role for R52 in Chromophore pK(a) Regulation in Photoactive Yellow Protein
Biochemistry, 45, 2006
|
|
1WQ8
 
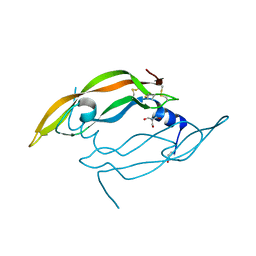 | | Crystal structure of Vammin, a VEGF-F from a snake venom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Vascular endothelial growth factor toxin | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
1WQ9
 
 | | Crystal structure of VR-1, a VEGF-F from a snake venom | | Descriptor: | Vascular endothelial growth factor | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-24 | | Release date: | 2004-12-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
2DDB
 
 | | Crystal structure of pseudecin from Pseudechis porphyriacus | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudecin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2DDA
 
 | | Crystal structure of pseudechetoxin from Pseudechis australis | | Descriptor: | FORMIC ACID, GLYCEROL, Pseudechetoxin, ... | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2006-01-25 | | Release date: | 2007-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2EPF
 
 | | Crystal Structure of Zinc-Bound Pseudecin From Pseudechis Porphyriacus | | Descriptor: | Pseudecin, SODIUM ION, ZINC ION | | Authors: | Suzuki, N, Yamazaki, Y, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2007-03-29 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of pseudechetoxin and pseudecin, two snake-venom cysteine-rich secretory proteins that target cyclic nucleotide-gated ion channels: implications for movement of the C-terminal cysteine-rich domain
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8WAG
 
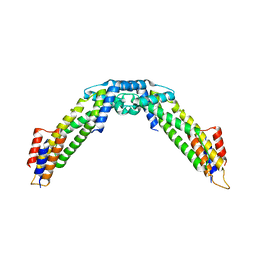 | | Crystal structure of the C-terminal fragment (residues 716-982) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Nakamura, Y, Takano, A, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
8WAF
 
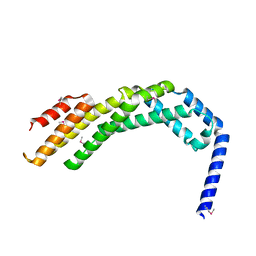 | | Crystal structure of the C-terminal fragment (residues 756-982 with the C864S mutation) of Arabidopsis thaliana CHUP1 | | Descriptor: | Protein CHUP1, chloroplastic | | Authors: | Shimada, A, Takano, A, Nakamura, Y, Kohda, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CHLOROPLAST UNUSUAL POSITIONING 1 is a plant-specific actin polymerization factor regulating chloroplast movement.
Plant Cell, 36, 2024
|
|
5GX9
 
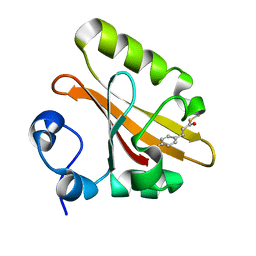 | | PYP mutant - E46Q | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Yonezawa, K. | | Deposit date: | 2016-09-16 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | NEUTRON DIFFRACTION (1.493 Å) | | Cite: | Neutron crystallography of photoactive yellow protein reveals unusual protonation state of Arg52 in the crystal
Sci Rep, 7, 2017
|
|
2ZOH
 
 | |
2ZOI
 
 | |
3VHZ
 
 | |
3VI0
 
 | |
7XWP
 
 | |
7XVY
 
 | | Human Estrogen Receptor beta Ligand-binding Domain in Complex with S-DPN | | Descriptor: | (2~{S})-2,3-bis(4-hydroxyphenyl)propanenitrile, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Furuya, N, Handa, C. | | Deposit date: | 2022-05-25 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Evaluating the correlation of binding affinities between isothermal titration calorimetry and fragment molecular orbital method of estrogen receptor beta with diarylpropionitrile (DPN) or DPN derivatives.
J.Steroid Biochem.Mol.Biol., 222, 2022
|
|
7XWQ
 
 | |
7XWR
 
 | |
7XVZ
 
 | |
