7XQC
 
 | |
6LWY
 
 | |
6LWZ
 
 | |
3HUG
 
 | |
3I7T
 
 | |
2O7G
 
 | |
2O8X
 
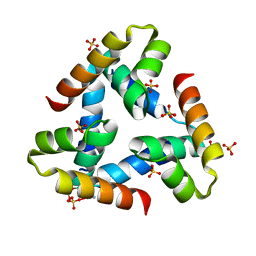 | |
5IQZ
 
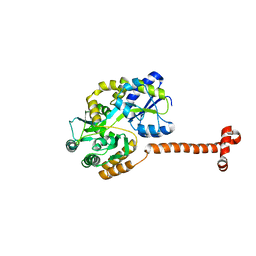 | | Crystal structure of N-terminal domain of Human SIRT7 | | Descriptor: | SIRT7 protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Thakur, K.G, Priyanka, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Crystal structure of the N-terminal domain of human SIRT7 reveals a three-helical domain architecture
Proteins, 84, 2016
|
|
5IFK
 
 | | Purine nucleoside phosphorylase | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Thakur, K.G, Priyanka, A. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Functional and Structural Characterization of Purine Nucleoside Phosphorylase from Kluyveromyces lactis and Its Potential Applications in Reducing Purine Content in Food
PLoS ONE, 11, 2016
|
|
6L3W
 
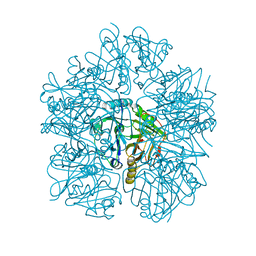 | | Crystal structure of BphC, a halotolerant catechol dioxygenase | | Descriptor: | Extra-diol dioxygenase BphC, FE (III) ION | | Authors: | Thakur, K.G, Solanki, V. | | Deposit date: | 2019-10-15 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Elucidation and Biochemical Characterization of Environmentally Relevant Novel Extradiol Dioxygenases Discovered by a Functional Metagenomics Approach.
mSystems, 4, 2019
|
|
5WZF
 
 | |
5WZ4
 
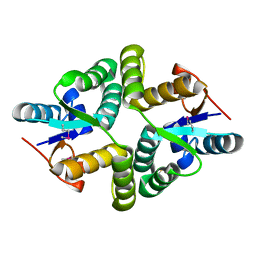 | |
4ILU
 
 | | Crystal structure of Mycobacterium tuberculosis CarD | | Descriptor: | RNA polymerase-binding transcription factor CarD, SULFATE ION | | Authors: | Thakur, K.G, Kaur, G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
7CT3
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C (MglC), SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-27 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 296, 2021
|
|
7CY1
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C, SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 2021
|
|
5ZFR
 
 | | Crystal structure of PilB, an extension ATPase motor of Type IV pilus, from Geobacter sulfurreducens | | Descriptor: | PHOSPHATE ION, Type IV pilus biogenesis ATPase PilB, ZINC ION | | Authors: | Thakur, K.G, Solanki, V, Kapoor, S. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of Type IVa pilus extension and retraction ATPase motors
FEBS J., 285, 2018
|
|
5ZFQ
 
 | |
8H5S
 
 | |
7YK3
 
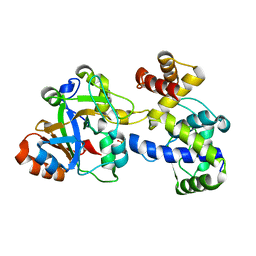 | | Crystal structure of DarTG toxin-antitoxin complex from Mycobacterium tuberculosis | | Descriptor: | DNA ADP-ribosyl glycohydrolase, DNA ADP-ribosyl transferase, PHOSPHATE ION | | Authors: | Deep, A, Kaur, J, Singh, L, Thakur, K.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into DarT toxin neutralization by cognate DarG antitoxin: ssDNA mimicry by DarG C-terminal domain keeps the DarT toxin inhibited.
Structure, 31, 2023
|
|
3GUZ
 
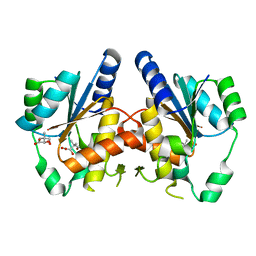 | | Structural and substrate-binding studies of pantothenate synthenate (PS)provide insights into homotropic inhibition by pantoate in PS's | | Descriptor: | PANTOATE, Pantothenate synthetase | | Authors: | Chakrabarti, K.S, Thakur, K.G, Gopal, B, Sarma, S.P. | | Deposit date: | 2009-03-30 | | Release date: | 2010-02-09 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and NMR studies of pantothenate synthetase provide insights into the mechanism of homotropic inhibition by pantoate
Febs J., 277, 2010
|
|
4MFR
 
 | | Crystal structure of Mycobacterium tuberculosis CarD | | Descriptor: | GLYCEROL, IODIDE ION, RNA polymerase-binding transcription factor CarD, ... | | Authors: | Kaur, G, Thakur, K.G. | | Deposit date: | 2013-08-28 | | Release date: | 2013-11-06 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Proteins, 82, 2014
|
|
4OZK
 
 | |
7C48
 
 | |
7C46
 
 | |
6A7V
 
 | | Crystal structure of Mycobacterium tuberculosis VapBC11 toxin-antitoxin complex | | Descriptor: | Antitoxin VapB11, PENTAETHYLENE GLYCOL, Ribonuclease VapC11, ... | | Authors: | Deep, A, Thakur, K.G. | | Deposit date: | 2018-07-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: targeting a tRNase to tackle mycobacterial adaptation.
Nucleic Acids Res., 46, 2018
|
|
