4FWT
 
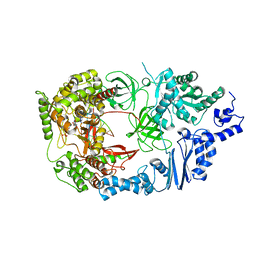 | | Complex structure of viral RNA polymerase form III | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-07-02 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
2DOK
 
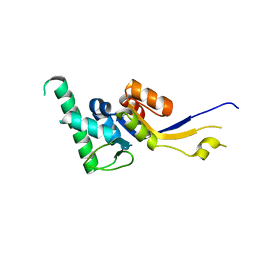 | | Crystal structure of the PIN domain of human EST1A | | Descriptor: | Telomerase-binding protein EST1A | | Authors: | Takeshita, D. | | Deposit date: | 2006-05-01 | | Release date: | 2007-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the PIN domain of human telomerase-associated protein EST1A
Proteins, 68, 2007
|
|
2EB1
 
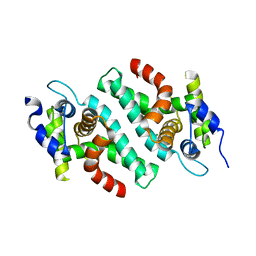 | | Crystal Structure of the C-Terminal RNase III Domain of Human Dicer | | Descriptor: | Endoribonuclease Dicer, MAGNESIUM ION | | Authors: | Takeshita, D, Zenno, S, Lee, W.C, Nagata, K, Saigo, K, Tanokura, M. | | Deposit date: | 2007-02-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homodimeric Structure and Double-stranded RNA Cleavage Activity of the C-terminal RNase III Domain of Human Dicer
J.Mol.Biol., 374, 2007
|
|
4O0L
 
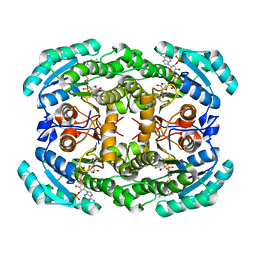 | |
4Q7J
 
 | | Complex structure of viral RNA polymerase | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu 1, ... | | Authors: | Takeshita, D. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular insights into replication initiation by Q beta replicase using ribosomal protein S1.
Nucleic Acids Res., 42, 2014
|
|
3AGP
 
 | | Structure of viral polymerase form I | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AGQ
 
 | | Structure of viral polymerase form II | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, LINKER, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AVT
 
 | | Structure of viral RNA polymerase complex 1 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVU
 
 | | Structure of viral RNA polymerase complex 2 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVW
 
 | | Structure of viral RNA polymerase complex 4 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVX
 
 | | Structure of viral RNA polymerase complex 5 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVV
 
 | | Structure of viral RNA polymerase complex 3 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.119 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVY
 
 | | Structure of viral RNA polymerase complex 6 | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VNV
 
 | | Complex structure of viral RNA polymerase II | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
3VNU
 
 | | Complex structure of viral RNA polymerase I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism for template-independent terminal adenylation activity of Q beta replicase
Structure, 20, 2012
|
|
8I5J
 
 | |
8I5K
 
 | |
3AK4
 
 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
3AQK
 
 | |
3AQN
 
 | |
3AQL
 
 | |
3AQM
 
 | | Structure of bacterial protein (form II) | | Descriptor: | MAGNESIUM ION, Poly(A) polymerase | | Authors: | Toh, Y, Takeshita, D, Tomita , K. | | Deposit date: | 2010-11-09 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Mechanism for the alteration of the substrate specificities of template-independent RNA polymerases
Structure, 19, 2011
|
|
3WFR
 
 | | tRNA processing enzyme complex 2 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFS
 
 | | tRNA processing enzyme complex 3 | | Descriptor: | Poly A polymerase, RNA (74-MER), SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.311 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFO
 
 | | tRNA processing enzyme (apo form 1) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
