5O98
 
 | | Binary complex of Catharanthus roseus Vitrosamine Synthase with NADP+ | | Descriptor: | Alcohol dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Stavrinides, A.K, Tatsis, E.C, Dang, T.T, Caputi, L, Stevenson, C.E.M, Lawson, D.M, Schneider, B, O'Connor, S.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a Short-Chain Dehydrogenase from Catharanthus roseus that Produces a New Monoterpene Indole Alkaloid.
Chembiochem, 19, 2018
|
|
4ZR6
 
 | | Lymnaea Stagnalis Acetylcholine Binding Protein in Complex with 3-[(4E)-4-[(3-methylimidazol-4-yl)methylene]-2,3-dihydropyrrol-5-yl]pyridine | | Descriptor: | 3-{(4E)-4-[(1-methyl-1H-imidazol-5-yl)methylidene]-3,4-dihydro-2H-pyrrol-5-yl}pyridine, Acetylcholine-binding protein | | Authors: | Bobango, J, Bryant, D, Denton, T.T, Talley, T.T. | | Deposit date: | 2015-05-11 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-guided design and analysis of arylidene anabaseines and myosmines
To be Published
|
|
1TFJ
 
 | | Crystal structure of Bovine Glycolipid transfer protein in complex with a fatty acid | | Descriptor: | CHLORIDE ION, DECANOIC ACID, GLYCEROL, ... | | Authors: | Airenne, T.T, Kidron, H, West, G, Nymalm, Y, Mattjus, P, Salminen, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural evidence for adaptive ligand binding of glycolipid transfer protein.
J.Mol.Biol., 355, 2006
|
|
4V3A
 
 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
2BH2
 
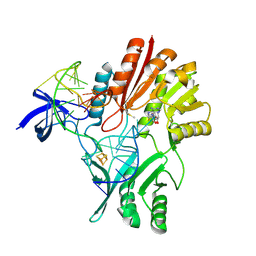 | | Crystal Structure of E. coli 5-methyluridine methyltransferase RumA in complex with ribosomal RNA substrate and S-adenosylhomocysteine. | | Descriptor: | 23S RIBOSOMAL RNA 1932-1968, 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2005-01-06 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function
Cell(Cambridge,Mass.), 120, 2005
|
|
2BV7
 
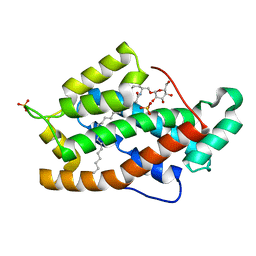 | | Crystal structure of GLTP with bound GM3 | | Descriptor: | GLYCOLIPID TRANSFER PROTEIN, N-{1-[(HEXOPYRANOSYLOXY)METHYL]-2-HYDROXYNONADECYL}TETRACOSANAMIDE, SULFATE ION | | Authors: | Kidron, H, Airenne, T.T, Nymalm, Y, Nylund, M, West, G, Mattjus, P, Salminen, T.A. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein.
J.Mol.Biol., 355, 2006
|
|
1ALI
 
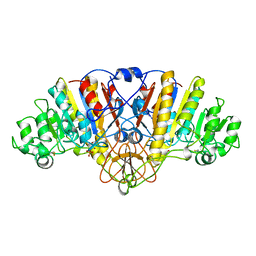 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1ALH
 
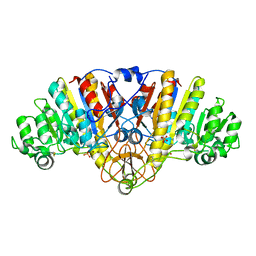 | | KINETICS AND CRYSTAL STRUCTURE OF A MUTANT E. COLI ALKALINE PHOSPHATASE (ASP-369-->ASN): A MECHANISM INVOLVING ONE ZINC PER ACTIVE SITE | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Tibbitts, T.T, Xu, X, Kantrowitz, E.R. | | Deposit date: | 1994-08-23 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics and crystal structure of a mutant Escherichia coli alkaline phosphatase (Asp-369-->Asn): a mechanism involving one zinc per active site.
Protein Sci., 3, 1994
|
|
1ALJ
 
 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1ANJ
 
 | | ALKALINE PHOSPHATASE (K328H) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
1ANI
 
 | | ALKALINE PHOSPHATASE (D153H, K328H) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Murphy, J.E, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-09-06 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations at positions 153 and 328 in Escherichia coli alkaline phosphatase provide insight towards the structure and function of mammalian and yeast alkaline phosphatases.
J.Mol.Biol., 253, 1995
|
|
1URA
 
 | | ALKALINE PHOSPHATASE (D51ZN) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Tibbitts, T.T, Murphy, J.E, Kantrowitz, E.R. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Kinetic and structural consequences of replacing the aspartate bridge by asparagine in the catalytic metal triad of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 257, 1996
|
|
1URB
 
 | | ALKALINE PHOSPHATASE (N51MG) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tibbitts, T.T, Murphy, J.E, Kantrowitz, E.R. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Kinetic and structural consequences of replacing the aspartate bridge by asparagine in the catalytic metal triad of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 257, 1996
|
|
1PU4
 
 | | Crystal structure of human vascular adhesion protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salminen, T.A, Airenne, T.T. | | Deposit date: | 2003-06-24 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human vascular adhesion protein-1: unique structural features with functional implications.
Protein Sci., 14, 2005
|
|
4V3N
 
 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
1GYR
 
 | | Mutant form of enoyl thioester reductase from Candida tropicalis | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, SULFATE ION | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-04-29 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
4W5H
 
 | |
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
1GUF
 
 | | Enoyl thioester reductase from Candida tropicalis | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH, B-SPECIFIC] 1, MITOCHONDRIAL, ... | | Authors: | Airenne, T.T, Torkko, J.M, Van Der Plas, S, Sormunen, R.T, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2002-01-25 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of Enoyl Thioester Reductase Involved in Mitochondrial Maintenance
J.Mol.Biol., 327, 2003
|
|
4W5J
 
 | |
7EW8
 
 | | Legionella pneumophila effector AnkD | | Descriptor: | ANK_REP_REGION domain-containing protein | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
4WV9
 
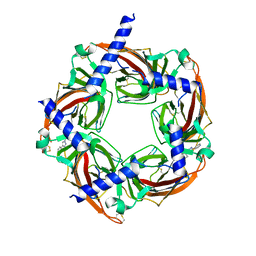 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane | | Descriptor: | (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J.M, Sankaran, B. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound.
To Be Published
|
|
4WUB
 
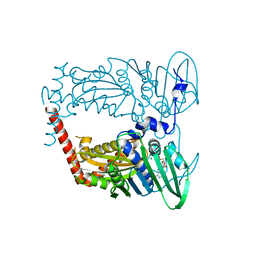 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
