5TII
 
 | |
8QO5
 
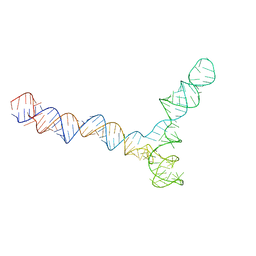 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 2024
|
|
8QO4
 
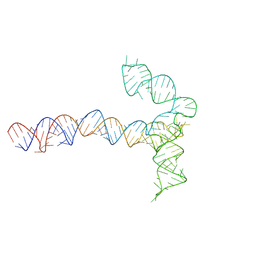 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 2024
|
|
8QO3
 
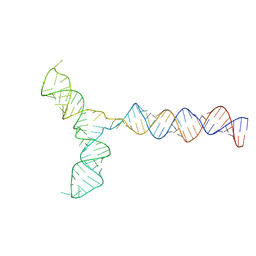 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 2024
|
|
8QO2
 
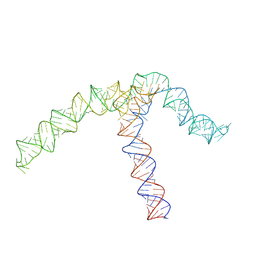 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 2024
|
|
4RGW
 
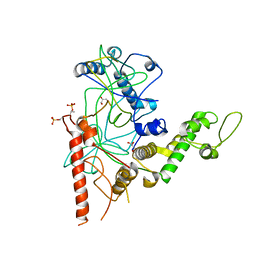 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | Descriptor: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | Authors: | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
4UAQ
 
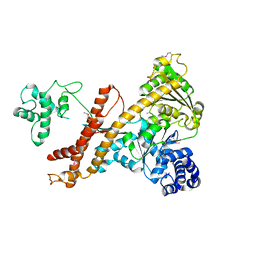 | | Crystal structure of the accessory translocation ATPase, SecA2, from Mycobacterium tuberculosis | | Descriptor: | Protein translocase subunit SecA 2 | | Authors: | Swanson-Smith, S, Ioerger, T.R, Rigel, N.W, Miller, B.K, Braunstein, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-09 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2.
J.Bacteriol., 198, 2015
|
|
4V69
 
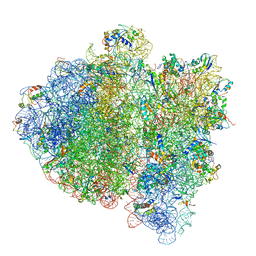 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7MX1
 
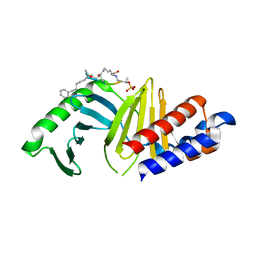 | | PLK-1 polo-box domain in complex with a high affinity macrocycle synthesized using a novel glutamic acid analog | | Descriptor: | ACE-PRO-LEU-ALA-SER-TPO, N-[(4S)-4,5-diamino-5-oxopentyl]-10-phenyldecanamide, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of a new orthogonally protected glutamic acid analog and its use in the preparation of high affinity polo-like kinase 1 polo-box domain - binding peptide macrocycles.
Org.Biomol.Chem., 19, 2021
|
|
1JTT
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | FORMIC ACID, Lysozyme, SODIUM ION, ... | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JTO
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | Lysozyme, Vh Single-Domain Antibody | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-21 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JTP
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | FORMIC ACID, LYSOZYME C, SODIUM ION, ... | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-21 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JUL
 
 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS IN A SECOND ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Knoechel, T.R, Hennig, M, Merz, A, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1996-05-03 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength.
J.Mol.Biol., 262, 1996
|
|
1JUK
 
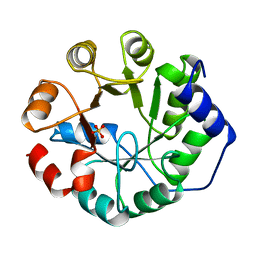 | | INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS IN A TRIGONAL CRYSTAL FORM | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, SULFATE ION | | Authors: | Knoechel, T.R, Hennig, M, Merz, A, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1996-05-03 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength.
J.Mol.Biol., 262, 1996
|
|
1MV2
 
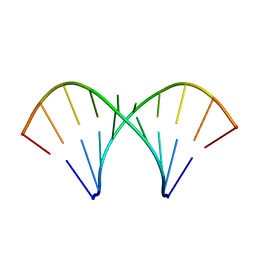 | | The tandem, Face-to-Face AP Pairs in 5'(rGGCAPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MV6
 
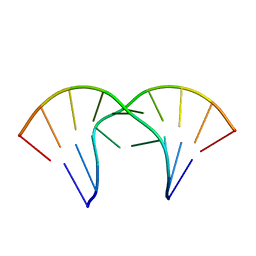 | | The tandem, Sheared PP Pairs in 5'(rGGCPPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MUV
 
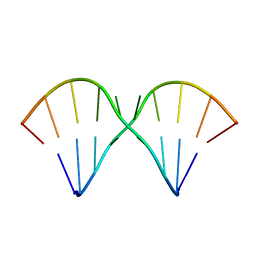 | | Sheared A(anti)-A(anti) Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of 5'(rGGCAAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Schroeder, S.J, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Sheared Aanti-Aanti Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of
5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MV1
 
 | | The Tandem, Sheared PA Pairs in 5'(rGGCPAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
8UPT
 
 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
8QQ3
 
 | | Streptavidin with a Ni-cofactor | | Descriptor: | 4-[4-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butylamino]-~{N}1,~{N}1'-di(quinolin-8-yl)cyclohexane-1,1-dicarboxamide, NICKEL (II) ION, Streptavidin | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial nickel chlorinase based on the biotin-streptavidin technology.
Chem.Commun.(Camb.), 60, 2024
|
|
8CW2
 
 | | Crystal structure of TDP1 complexed with compound XZ760 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-({(4R)-7-phenyl-2-[4-(2-{[4-(pyridin-2-yl)phenyl]methoxy}ethyl)phenyl]imidazo[1,2-a]pyridin-3-yl}amino)benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification.
Rsc Chem Biol, 4, 2023
|
|
8CVQ
 
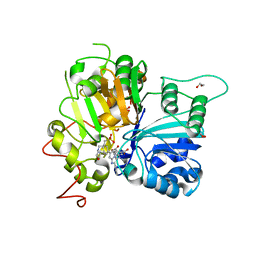 | | Crystal structure of TDP1 complexed with compound XZ761 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification.
Rsc Chem Biol, 4, 2023
|
|
3V39
 
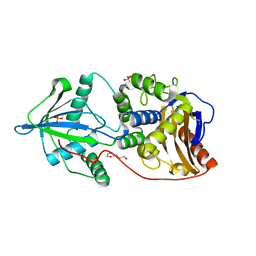 | | Bd3459, A Predatory Peptidoglycan Endopeptidase from Bdellovibrio bacteriovorus | | Descriptor: | 2-AMINOETHANESULFONIC ACID, D-alanyl-D-alanine carboxypeptidase, SULFATE ION, ... | | Authors: | Lovering, A.L, Lerner, T.R, Sockett, R.E. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness.
Plos Pathog., 8, 2012
|
|
6MJ5
 
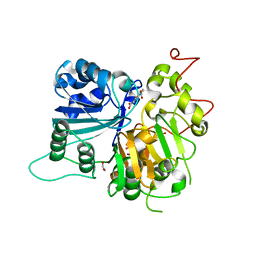 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ519 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-nitroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6N19
 
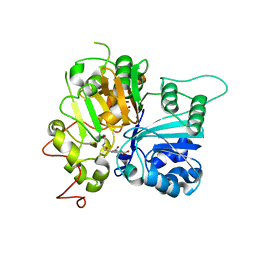 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-carboxybutanoyl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
