3UDE
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1B | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UD5
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1A | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UDV
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1C | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
4F7V
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1D (HP26) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Bisubstrate analog inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New lead exhibits a distinct binding mode.
Bioorg.Med.Chem., 20, 2012
|
|
2NUG
 
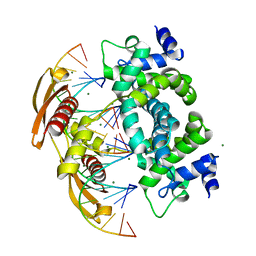 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 1.7-Angstrom Resolution | | Descriptor: | 5'-R(P*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*G)-3', 5'-R(P*AP*GP*UP*GP*GP*CP*CP*UP*UP*GP*C)-3', MAGNESIUM ION, ... | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
2NUF
 
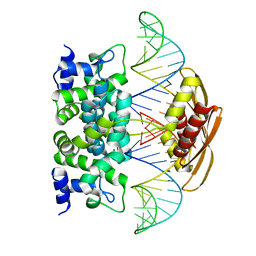 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.5-Angstrom Resolution | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
2NUE
 
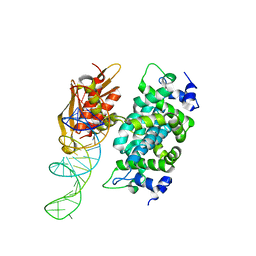 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.9-Angstrom Resolution | | Descriptor: | 46-MER, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
2PCX
 
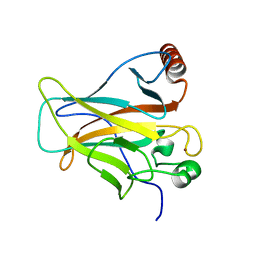 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
6AN4
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-39 (J1F) | | Descriptor: | ((2-(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carboxamido)-N-(2-((((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)amino)-2-oxoethyl)acetamido)methyl)phosphonic acid, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bisubstrate analog inhibitors of HPPK: Transition state mimetics
to be published
|
|
6AN6
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-72 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-{2-[{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}(phosphonomethyl)amino]ethyl}-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bisubstrate analogue inhibitors of HPPK: Transition state mimetics
to be published
|
|
4PZV
 
 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with bisubstrate analog HPPK inhibitor J1D | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK-DHPS from the biowarfare agent Francisella tularensis.
Febs J., 281, 2014
|
|
7KDR
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]sulfonyl}-5'-deoxyadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.488 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
7KDO
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-73 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
1BAK
 
 | |
2QX0
 
 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2007-08-10 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1VVE
 
 | |
1VVD
 
 | |
1VVC
 
 | |
