7OQH
 
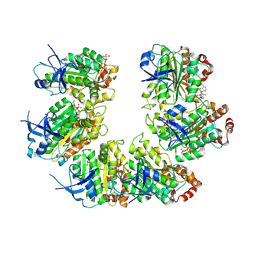 | | CryoEM structure of the transcription termination factor Rho from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Saridakis, E, Vishwakarma, R, Lai Kee Him, J, Martin, K, Simon, I, Cohen-Gonsaud, M, Coste, F, Bron, P, Margeat, E, Boudvillain, M. | | Deposit date: | 2021-06-03 | | Release date: | 2022-02-09 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism.
Commun Biol, 5, 2022
|
|
3EUN
 
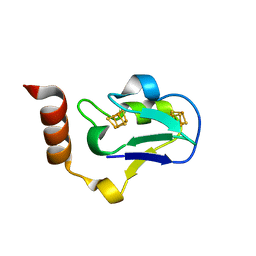 | |
6SHG
 
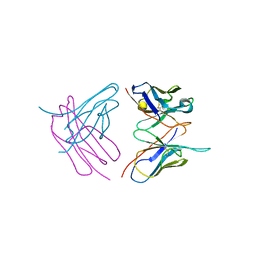 | | Diffraction data for RoAb13 crystal co-crystallised with PIYDIN and its RoAb13 structure | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chain, B, Morgan, M, Kassen, S.C, Chayen, N.E. | | Deposit date: | 2019-08-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.35050821 Å) | | Cite: | X-ray crystallography based model of the antibody RoAb13 bound to peptidomimetics of the HIV receptor chemokine receptor CCR5
To Be Published
|
|
5J6S
 
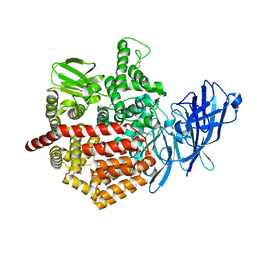 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | Descriptor: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | Deposit date: | 2016-04-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
5K1V
 
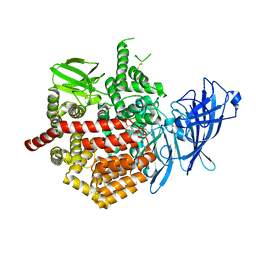 | | Crystal structure of Endoplasmic Reticulum aminopeptidase 2 (ERAP2) in complex with a diaminobenzoic acid derivative ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Papakyriakou, A, Giastas, P, Mpakali, A, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
7NW3
 
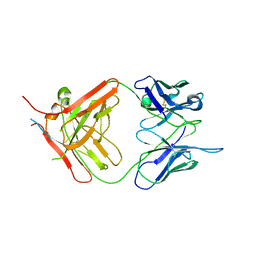 | | X-ray crystallographic study of PIYDIN, which contains the truncation determinants of binding PI and N, bound to RoAb13, a CCR5 antibody | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Saridakis, E, Helliwell, J.R, Govada, L, Chayen, N.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.200011 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
5NHI
 
 | |
4JBS
 
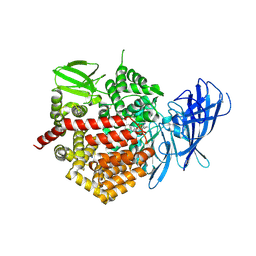 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 in complex with PHOSPHINIC PSEUDOTRIPEPTIDE inhibitor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Birtley, J, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3EXY
 
 | |
5CU5
 
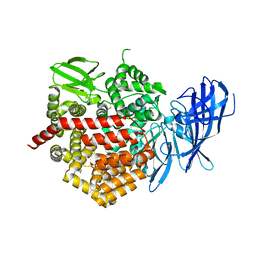 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
4IP1
 
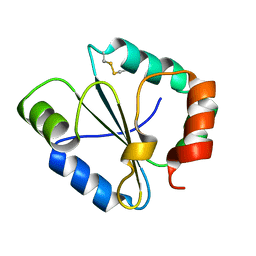 | |
4IP6
 
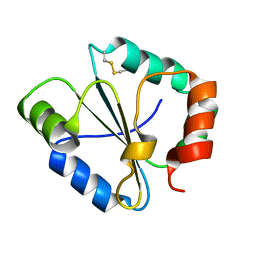 | |
6YDX
 
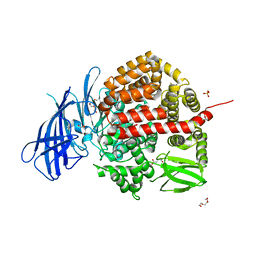 | | Insulin-regulated aminopeptidase complexed with a macrocyclic peptidic inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[[[(4~{R},8~{S},11~{S})-11-azanyl-8-[(4-hydroxyphenyl)methyl]-6,10-bis(oxidanylidene)-1,2-dithia-5,9-diazacyclotridec-4-yl]carbonylamino]methyl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Saridakis, E, Giastas, P, Stratikos, E. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
4S2S
 
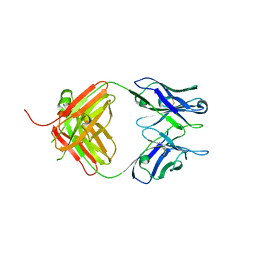 | | Crystal Structure of Fab fragment of monoclonal antibody RoAb13 | | Descriptor: | RoAb13 Fab Heavy chain, RoAb13 Fab Light chain | | Authors: | Chain, B, Arnold, J, Akthar, S, Noursadeghi, M, Lapp, T, Ji, C, Naider, D, Zhang, Y, Govada, L, Saridakis, E, Chayen, N.E. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Linear Epitope in the N-Terminal Domain of CCR5 and Its Interaction with Antibody.
Plos One, 10, 2015
|
|
4E36
 
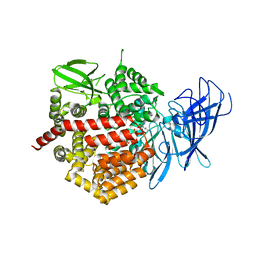 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 variant N392K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Saridakis, E, Pegias, P, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing.
J.Immunol., 189, 2012
|
|
5MJ6
 
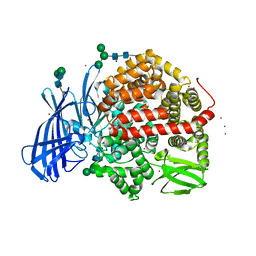 | | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Mpakali, A, Stratikos, E, Saridakis, E, Giastas, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity.
J. Med. Chem., 60, 2017
|
|
3SE6
 
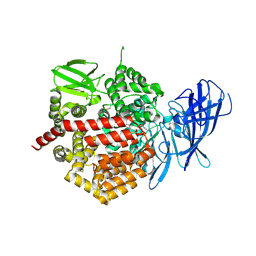 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Saridakis, E, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | The crystal structure of human endoplasmic reticulum aminopeptidase 2 reveals the atomic basis for distinct roles in antigen processing.
Biochemistry, 51, 2012
|
|
7NJZ
 
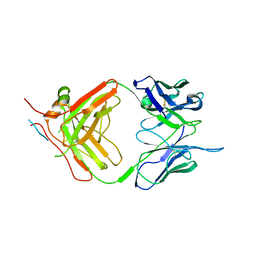 | | X-ray crystallography study of RoAb13 which binds to PIYDIN, a part of the CCR5 N terminal domain | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Helliwell, J.R, Chayen, N, Saridakis, E, Govada, L. | | Deposit date: | 2021-02-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
3PFU
 
 | | N-terminal domain of Thiol:disulfide interchange protein DsbD in its reduced form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thiol:disulfide interchange protein dsbD | | Authors: | Mavridou, D.A.I, Saridakis, E, Ferguson, S.J, Redfield, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxidation state-dependent protein-protein interactions in disulfide cascades
J.Biol.Chem., 286, 2011
|
|
4Z7I
 
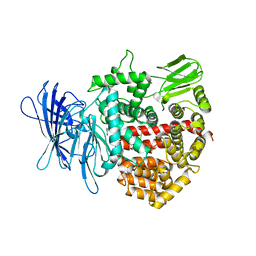 | | Crystal structure of insulin regulated aminopeptidase in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DG025 transition-state analogue enzyme inhibitor, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-04-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
5AB0
 
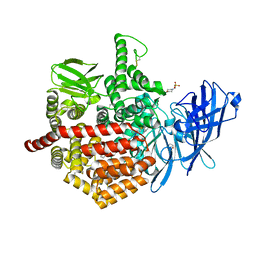 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
5AB2
 
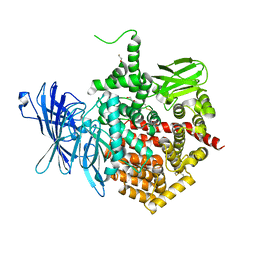 | | Crystal structure of aminopeptidase ERAP2 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Saridakis, E, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-31 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (Er) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
5C97
 
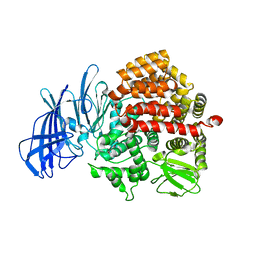 | | Insulin regulated aminopeptidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucyl-cystinyl aminopeptidase, ... | | Authors: | Mpakali, A, Saridakis, E, Harlos, K, Zhao, Y, Stratikos, E. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Crystal Structure of Insulin-Regulated Aminopeptidase with Bound Substrate Analogue Provides Insight on Antigenic Epitope Precursor Recognition and Processing.
J Immunol., 195, 2015
|
|
2ZVS
 
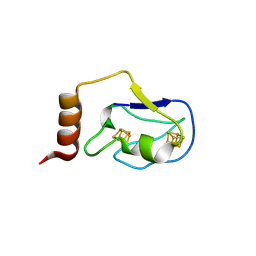 | | Crystal structure of the 2[4FE-4S] ferredoxin from escherichia coli | | Descriptor: | IRON/SULFUR CLUSTER, Uncharacterized ferredoxin-like protein yfhL | | Authors: | Giastas, P, Mavridis, M.I. | | Deposit date: | 2008-11-18 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insight into the protein and solvent contributions to the reduction potentials of [4Fe-4S]2+/+ clusters: crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin
J.Biol.Inorg.Chem., 14, 2009
|
|
