1ZTB
 
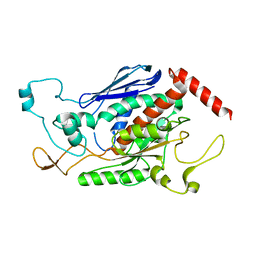 | | Crystal Structure of Chorismate Synthase from Mycobacterium tuberculosis | | Descriptor: | Chorismate synthase | | Authors: | Dias, M.V.B, Borges, J.C, Ely, F, Pereira, J.H, Canduri, F, Ramos, C.H.I, Frazzon, J, Palma, M.S, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2005-05-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of chorismate synthase from Mycobacterium tuberculosis
J.Struct.Biol., 154, 2006
|
|
6D6X
 
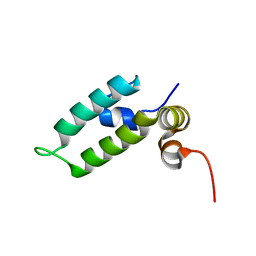 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
8EOD
 
 | |
6WIU
 
 | |
6BBA
 
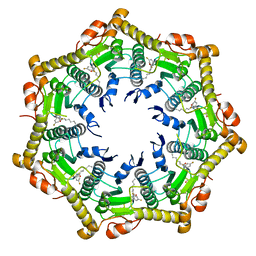 | |
6N99
 
 | | Xylose isomerase 2F1 variant from Streptomyces sp. F-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6N98
 
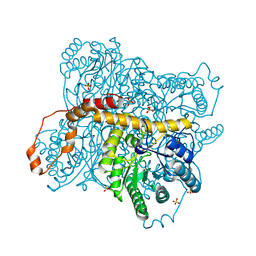 | | Xylose isomerase 1F1 variant from Streptomyces sp. F-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, Xylose isomerase | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6NB1
 
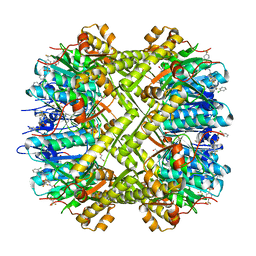 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
6NAH
 
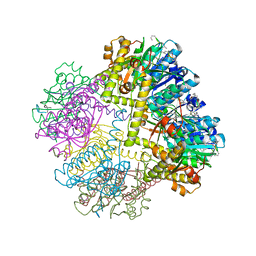 | |
6NAW
 
 | |
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
