3FLM
 
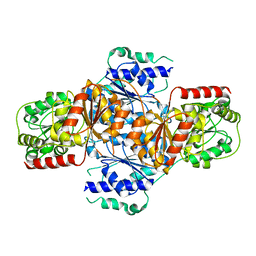 | | Crystal structure of menD from E.coli | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Priyadarshi, A, Hwang, K.Y. | | Deposit date: | 2008-12-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights of the MenD from Escherichia coli reveal ThDP affinity.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3IHC
 
 | |
3IIG
 
 | |
3ILB
 
 | |
3IHD
 
 | |
3IHF
 
 | |
3IIH
 
 | |
3IHE
 
 | |
3ILC
 
 | |
3GNT
 
 | |
3GNS
 
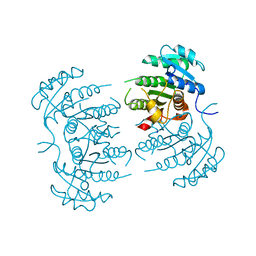 | |
3GR6
 
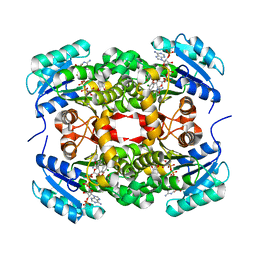 | |
3HWX
 
 | |
3HWW
 
 | |
4S0P
 
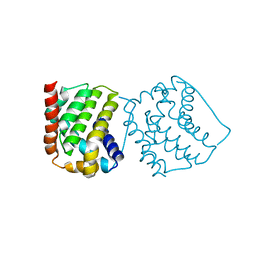 | |
4S0O
 
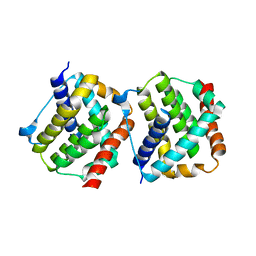 | |
3E6E
 
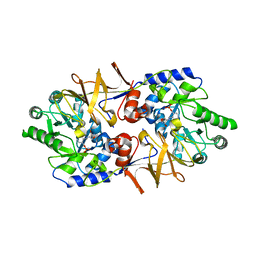 | | Crystal structure of Alanine racemase from E.faecalis complex with cycloserine | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3E5P
 
 | | Crystal structure of alanine racemase from E.faecalis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine racemase, NONAETHYLENE GLYCOL, ... | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
2NX8
 
 | |
3H17
 
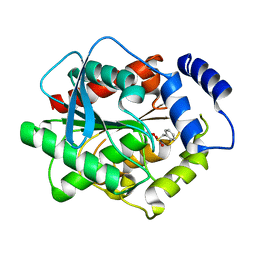 | | Crystal structure of EstE5-PMSF (I) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3H18
 
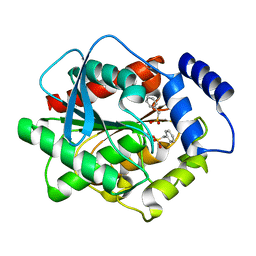 | | Crystal structure of EstE5-PMSF (II) | | Descriptor: | Esterase/lipase, phenylmethanesulfonic acid | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2009-04-11 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3CMD
 
 | | Crystal structure of peptide deformylase from VRE-E.faecium | | Descriptor: | FE (III) ION, MALONATE ION, Peptide deformylase, ... | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2008-03-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into the antibacterial drug design and architectural mechanism of peptide recognition from the E. faecium peptide deformylase structure.
Proteins, 74, 2009
|
|
3DNM
 
 | |
3FAK
 
 | |
3GVY
 
 | |
