4DEL
 
 | | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from M. tuberculosis | | Descriptor: | ACETATE ION, Fructose-bisphosphate aldolase, HEXAETHYLENE GLYCOL, ... | | Authors: | Pegan, S.D, Mesecar, A.D. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
3OX2
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | 2-hydroxy-8,9-dimethoxy-6H-isoindolo[2,1-a]indol-6-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OWX
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | 2-[4-(furan-2-ylcarbonyl)piperazin-1-yl]-6,7-dimethoxyquinazolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-20 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OX3
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(2-methoxy-1H-dipyrido[2,3-a:3',2'-e]pyrrolizin-11-yl)ethyl]furan-2-carboxamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OVM
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OX1
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-{2-[7-(methylsulfamoyl)naphthalen-1-yl]ethyl}acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3OWH
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
4P61
 
 | |
4DEF
 
 | | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from M. tuberculosis | | Descriptor: | ACETATE ION, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Capodagli, G.C, Pegan, S.D. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
4TO8
 
 | | Methicillin-Resistant Staphylococcus Aureus Class IIb Fructose 1,6-Bisphosphate Aldolase | | Descriptor: | CITRATE ANION, Fructose-1,6-bisphosphate aldolase, class II, ... | | Authors: | Capodagli, G.C, Pegan, S.D. | | Deposit date: | 2014-06-05 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Methicillin-Resistant Staphylococcus aureus's Class IIb Fructose 1,6-Bisphosphate Aldolase.
Biochemistry, 53, 2014
|
|
8DCY
 
 | | CCHFV GP38 Hoti/Kosovo bound with 13G8 Fab | | Descriptor: | 13G8 Heavy Chain, 13G8 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Durie, I.A, Bergeron, E, Pegan, S.D, McGuire, J. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural characterization of protective non-neutralizing antibodies targeting Crimean-Congo hemorrhagic fever virus.
Nat Commun, 13, 2022
|
|
8DDK
 
 | | CCHFV GP38 Hoti/Kosovo bound with CC5_17 | | Descriptor: | Envelopment polyprotein, Heavy Chain CC5-17, Light Chain CC5_17, ... | | Authors: | Durie, I.A, Bergeron, E, Pegan, S.D, McGuire, J. | | Deposit date: | 2022-06-18 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.86 Å) | | Cite: | Structural characterization of protective non-neutralizing antibodies targeting Crimean-Congo hemorrhagic fever virus.
Nat Commun, 13, 2022
|
|
8DC5
 
 | | CCHFV GP38 Hoti/Kosovo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, IODIDE ION | | Authors: | Durie, I.A, Bergeron, E, Pegan, S.D, McGuire, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural characterization of protective non-neutralizing antibodies targeting Crimean-Congo hemorrhagic fever virus.
Nat Commun, 13, 2022
|
|
7JMS
 
 | | Structure of the Hazara virus OTU bound to ubiquitin | | Descriptor: | CALCIUM ION, GLYCEROL, Polyubiquitin-B, ... | | Authors: | Dzimianski, J.V, Pegan, S.D. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Flipping the substrate preference of Hazara virus ovarian tumour domain protease through structure-based mutagenesis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8GH7
 
 | | 142D6 bound to BIR3-XIAP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BIR3 inhibitor MAA-CHG-PRO-ZHW, E3 ubiquitin-protein ligase XIAP, ... | | Authors: | Garza-Granados, A, McGuire, J, Baggio, C, Pellecchia, M, Pegan, S.D. | | Deposit date: | 2023-03-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a Potent and Orally Bioavailable Lys-Covalent Inhibitor of Apoptosis Protein (IAP) Antagonist.
J.Med.Chem., 66, 2023
|
|
6MDH
 
 | | X-ray crystal structure of ISG15 from Myotis davidii | | Descriptor: | Ubiquitin-like protein ISG15 | | Authors: | Goodwin, O.Y, Langley, C.A, Dzimianski, J.V, Daczkowski, C.M, Pegan, S.D. | | Deposit date: | 2018-09-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of interferon-stimulated gene product 15 (ISG15) from the bat species Myotis davidii and the impact of interdomain ISG15 interactions on viral protein engagement.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4HXD
 
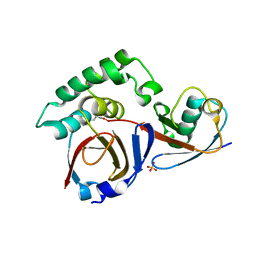 | |
5JZE
 
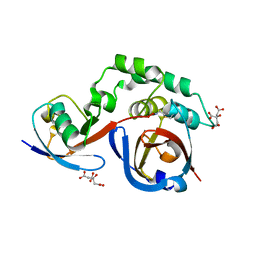 | | Erve virus viral OTU domain protease in complex with mouse ISG15 | | Descriptor: | CITRATE ANION, RNA-dependent RNA polymerase, Ubiquitin-like protein ISG15, ... | | Authors: | Deaton, M.K, Dzimianski, J.V, Pegan, S.D. | | Deposit date: | 2016-05-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Biochemical and Structural Insights into the Preference of Nairoviral DeISGylases for Interferon-Stimulated Gene Product 15 Originating from Certain Species.
J.Virol., 90, 2016
|
|
3MB2
 
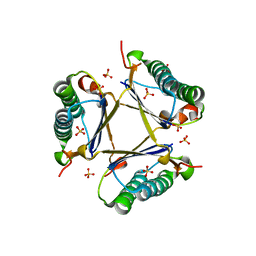 | | Kinetic and Structural Characterization of a Heterohexamer 4-Oxalocrotonate Tautomerase from Chloroflexus aurantiacus J-10-fl: Implications for Functional and Structural Diversity in the Tautomerase Superfamily | | Descriptor: | 4-oxalocrotonate tautomerase family enzyme - alpha subunit, 4-oxalocrotonate tautomerase family enzyme - beta subunit, SULFATE ION | | Authors: | Burks, E.A, Fleming, C.D, Mesecar, A.D, Whitman, C.P, Pegan, S.D. | | Deposit date: | 2010-03-24 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily
Biochemistry, 49, 2010
|
|
7MC9
 
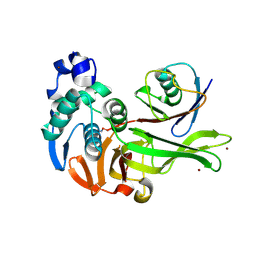 | | X-RAY STRUCTURE OF PEDV PAPAIN-LIKE PROTEASE 2 bound to UB-PA | | Descriptor: | 3C-like proteinase, Ubiquitin, ZINC ION, ... | | Authors: | Durie, I.A, Dzimianski, J.V, Daczkowski, C.M, Pegan, S.D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural insights into the interaction of papain-like protease 2 from the alphacoronavirus porcine epidemic diarrhea virus and ubiquitin
Acta Cryst. D, 77, 2021
|
|
7SBI
 
 | |
7SKR
 
 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor 37 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(2-methoxypyridin-4-yl)methyl]-2-[(1R)-1-(naphthalen-1-yl)ethyl]-2-azaspiro[3.3]heptane-6-carboxamide, ... | | Authors: | Durie, I, Shepard, J, Freitas, B, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
7SKQ
 
 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor GRL-0617 | | Descriptor: | 3C-like proteinase, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ZINC ION | | Authors: | Freitas, B, Durie, I, Shepard, J, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
3TAO
 
 | | Structure of Mycobacterium tuberculosis triosephosphate isomerase bound to phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Connor, S.E, Capodagli, G.C, Deaton, M.K, Pegan, S.D. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3TA6
 
 | | Structure of Mycobacterium tuberculosis triosephosphate isomerase | | Descriptor: | CITRATE ANION, Triosephosphate isomerase | | Authors: | Connor, S.E, Capodagli, G.C, Deaton, M.K, Pegan, S.D. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
