1Y64
 
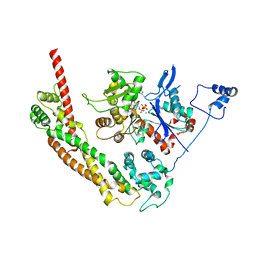 | | Bni1p Formin Homology 2 Domain complexed with ATP-actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Otomo, T, Tomchick, D.R, Otomo, C, Panchal, S.C, Machius, M, Rosen, M.K. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain
Nature, 433, 2005
|
|
6E6J
 
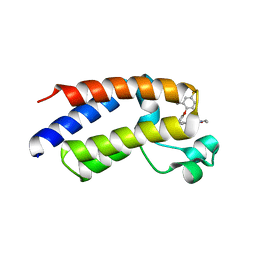 | | BRD2_Bromodomain2 complex with inhibitor 744 | | Descriptor: | Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Park, C.H, Bigelow, L. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
5K0M
 
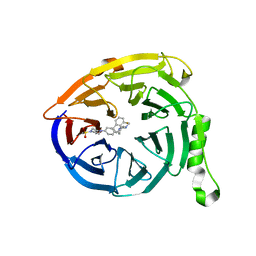 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
6X81
 
 | |
6X85
 
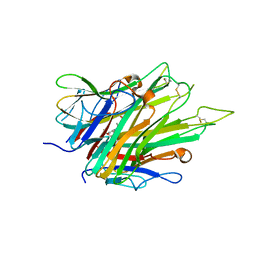 | | Crystal Structure of TNFalpha with indolinone compound 9 | | Descriptor: | 1-{[2-(difluoromethoxy)phenyl]methyl}-2,2-dimethyl-1,2-dihydro-3H-indol-3-one, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
8USS
 
 | | IL17A complexed to Compound 7 | | Descriptor: | 4,5-dichloro-N-[(1S)-1-cyclohexyl-2-{[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]amino}-2-oxoethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, Interleukin-17A | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 2024
|
|
8USR
 
 | | IL17A homodimer complexed to Compound 23 | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1-[[(1~{S})-1-(8~{a}~{H}-imidazo[1,2-a]pyrimidin-2-yl)ethyl]amino]-1-oxidanylidene-4-phenyl-butan-2-yl]-4,5-bis(chloranyl)-1~{H}-pyrrole-2-carboxamide | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 2024
|
|
8DYG
 
 | | IL17A homodimer bound to Compound 7 | | Descriptor: | (5P)-2-hydroxy-5-(6-methylquinolin-5-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYH
 
 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYI
 
 | | IL17A homodimer bound to Compound 5 | | Descriptor: | (5P)-5-[5-(benzylamino)pyridin-3-yl]-N-[2-(morpholin-4-yl)ethyl]-1H-indazol-3-amine, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYF
 
 | | IL17A homodimer bound to Compound 10 | | Descriptor: | (5M)-3-[({2-[2-(2-{2-[2-({[(5M)-3-carboxy-5-(5,8-dihydroquinolin-4-yl)phenyl]amino}methyl)phenoxy]ethoxy}ethoxy)ethoxy]phenyl}methyl)amino]-5-(quinolin-4-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
7SIU
 
 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
6X83
 
 | |
6X82
 
 | | Crystal Structure of TNFalpha with isoquinoline compound 4 | | Descriptor: | 8-[4-(2-{5-[(4-methylpiperazin-1-yl)methyl]-2-(1H-pyrrolo[3,2-c]pyridin-3-yl)phenoxy}ethyl)phenyl]isoquinoline, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
6X86
 
 | | Crystal Structure of TNFalpha with indolinone compound 11 | | Descriptor: | 3-[(6-{2-[(3R)-4-(hydroxyacetyl)-3-methylpiperazin-1-yl]pyrimidin-5-yl}-2,2-dimethyl-3-oxo-2,3-dihydro-1H-indol-1-yl)methyl]pyridine-2-carbonitrile, Tumor necrosis factor | | Authors: | Longenecker, K.L, Stoll, V.S. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Development of Orally Efficacious Allosteric Inhibitors of TNF alpha via Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
6NCN
 
 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-(3-chlorophenyl)cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
6NCO
 
 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-[5-chloro-4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
6ONY
 
 | | BRD2_Bromodomain1 complex with inhibitor 744 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Bigelow, L. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
5UEW
 
 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEY
 
 | | BRD4_BD2_A-1412838 | | Descriptor: | 5-[2-(2,4-difluorophenoxy)-5-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}phenyl]-4-ethoxy-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEX
 
 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEU
 
 | | BRD4_BD2_A-1107604 | | Descriptor: | Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UF0
 
 | | BRD4_BD2-A-35165 | | Descriptor: | 2-methyl-5-(methylamino)-6-phenylpyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEZ
 
 | | BRD4_BD2_A-1342843 | | Descriptor: | 5-methoxy-2-methyl-6-(2-phenoxyphenyl)pyridazin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5WI0
 
 | |
