3GHY
 
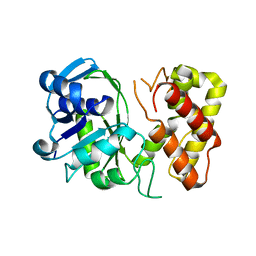 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
3GFO
 
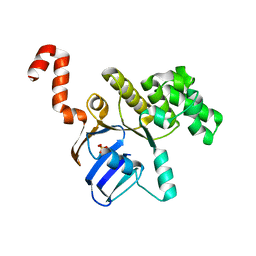 | | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ. | | Descriptor: | Cobalt import ATP-binding protein cbiO 1, SULFATE ION | | Authors: | Ramagopal, U.A, Morano, C, Toro, R, Dickey, M, Do, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of cbiO1 from clostridium perfringens: Part of the ABC transporter complex cbiONQ
To be published
|
|
3G8R
 
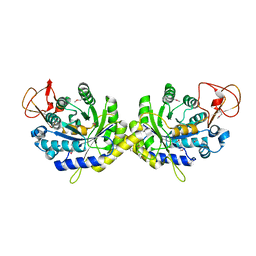 | | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472 | | Descriptor: | Probable spore coat polysaccharide biosynthesis protein E, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472
To be Published
|
|
3GG9
 
 | | CRYSTAL STRUCTURE OF putative D-3-phosphoglycerate dehydrogenase oxidoreductase from Ralstonia solanacearum | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Putative D-3-Phosphoglycerate Dehydrogenase from Ralstonia Solanacearum
To be Published
|
|
3GPI
 
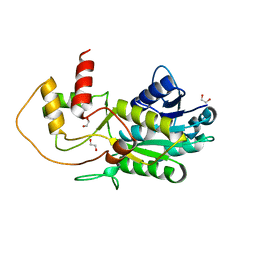 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
3GRZ
 
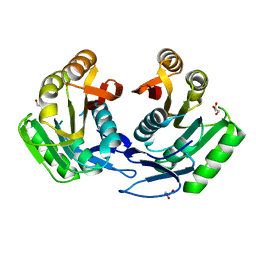 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3GY1
 
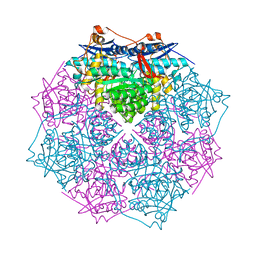 | | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium beijerinckii NCIMB 8052 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium
beijerinckii NCIMB 8052
To be Published
|
|
3GZ4
 
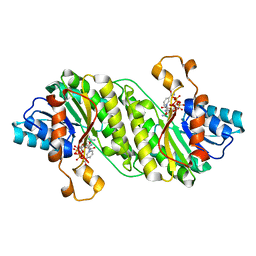 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
3H12
 
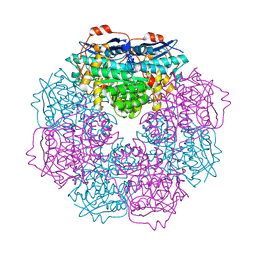 | | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50 | | Descriptor: | SODIUM ION, mandelate racemase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative mandelate racemase from Bordetella Bronchiseptica RB50
To be Published
|
|
3GT7
 
 | | CRYSTAL STRUCTURE OF SIGNAL RECEIVER DOMAIN OF SIGNAL TRANSDUCTION HISTIDINE KINASE FROM Syntrophus aciditrophicus | | Descriptor: | Sensor protein | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Signal Transduction Kinase from Syntrophus Aciditrophicus
To be Published
|
|
3GP4
 
 | | Crystal structure of putative MerR family transcriptional regulator from listeria monocytogenes | | Descriptor: | D-METHIONINE, GLYCEROL, SODIUM ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of putative MerR family transcriptional regulator from Listeria monocytogenes
To be published
|
|
3GMF
 
 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3H7A
 
 | | CRYSTAL STRUCTURE OF SHORT-CHAIN DEHYDROGENASE FROM Rhodopseudomonas palustris | | Descriptor: | UNKNOWN LIGAND, short chain dehydrogenase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CRYSTAL STRUCTURE OF SHORT-CHAIN DEHYDROGENASE FROM Rhodopseudomonas palustris
To be Published
|
|
3H7L
 
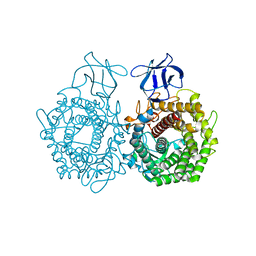 | | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus | | Descriptor: | ENDOGLUCANASE, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-27 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ENDOGLUCANASE-RELATED PROTEIN FROM Vibrio parahaemolyticus
To be Published
|
|
3GY0
 
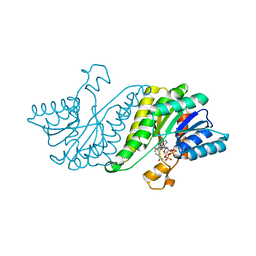 | | Crystal structure of putatitve short chain dehydrogenase FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putatitve short chain dehydrogenase
FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP
To be Published
|
|
3GMS
 
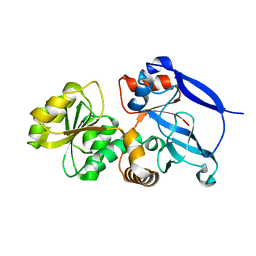 | |
3HCW
 
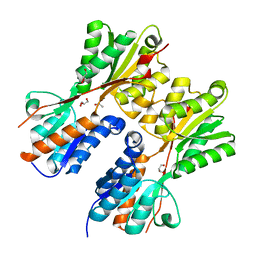 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | Descriptor: | GLYCEROL, Maltose operon transcriptional repressor | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
3HG7
 
 | | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449
To be Published
|
|
3HJG
 
 | | Crystal structure of putative alpha-ribazole-5'-phosphate phosphatase CobC FROM Vibrio parahaemolyticus | | Descriptor: | Putative alpha-ribazole-5'-phosphate phosphatase CobC, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alpha-ribazole-5' -phosphate phosphatase FROM Vibrio parahaemolyticus
To be Published
|
|
3HIN
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009 | | Descriptor: | Putative 3-hydroxybutyryl-CoA dehydratase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009
To be Published
|
|
3HN2
 
 | | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens GS-15 | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens
To be Published
|
|
3I42
 
 | |
3I47
 
 | | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Enoyl CoA hydratase/isomerase (Crotonase) | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila
subsp. pneumophila str. Philadelphia 1
To be Published
|
|
3HU5
 
 | | CRYSTAL STRUCTURE OF isochorismatase family protein from Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | | Descriptor: | Isochorismatase family protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-12 | | Release date: | 2009-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF isochorismatase family protein from Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough
To be Published
|
|
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
