1APF
 
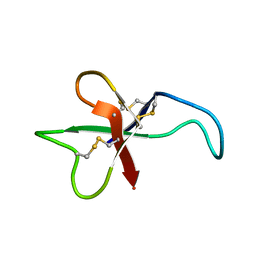 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
1RON
 
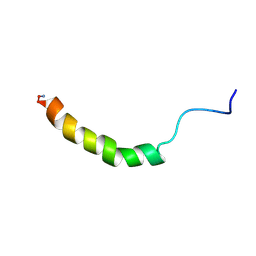 | |
1AHL
 
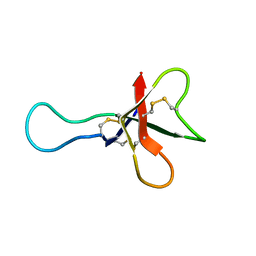 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-A | | Authors: | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | Deposit date: | 1994-10-28 | | Release date: | 1995-11-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
1ZEC
 
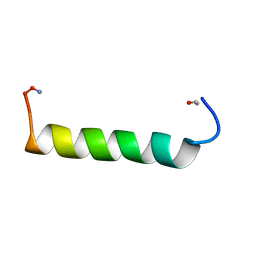 | | NMR Solution structure of NEF1-25, 20 structures | | Descriptor: | NEF1-25 | | Authors: | Barnham, K.J, Monks, S.A, Hinds, M.G, Azad, A.A, Norton, R.S. | | Deposit date: | 1996-12-18 | | Release date: | 1998-01-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Biochemistry, 36, 1997
|
|
7MYR
 
 | | BACE-1 in complex with compound #18 | | Descriptor: | (4aR,7aR)-6-(5-fluoropyrimidin-2-yl)-7a-(1,2-thiazol-5-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
7MYU
 
 | | BACE-1 in complex with compound #22 | | Descriptor: | Beta-secretase 1, N-{3-[(4aR,7aS)-2-amino-6-(5-fluoropyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-4-fluorophenyl}-5-methoxypyrazine-2-carboxamide, SULFATE ION | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
7MYI
 
 | | BACE-1 in complex with compound #6 | | Descriptor: | (4aR,7aR)-6-(pyrimidin-2-yl)-7a-(thiophen-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
6UWP
 
 | | BACE-1 in complex with compound #32 | | Descriptor: | (1R,2R)-2-[(4aR,7aR)-2-amino-6-(pyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVP
 
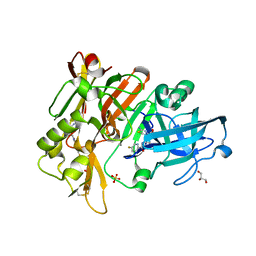 | | BACE-1 in complex with compound #3 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]cyclopropyl}-5-fluoropyridine-2-carboxamide, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UWV
 
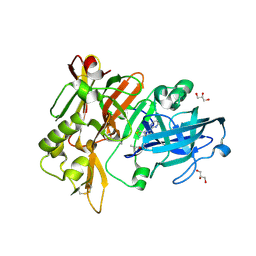 | | BACE-1 in complex with compound #34 | | Descriptor: | (4aR,7aR)-7a-[(1R,2R)-2-(2-{[(1R,2R)-2-methylcyclopropyl]methoxy}propan-2-yl)cyclopropyl]-6-(pyrimidin-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Stout, S.L. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVY
 
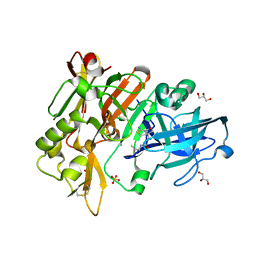 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UVV
 
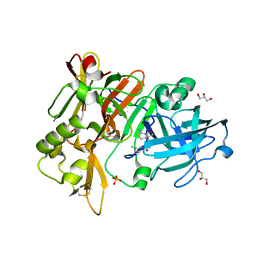 | | BACE-1 in complex with compound #17 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-butylcyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
4X7I
 
 | | Crystal Structure of BACE with amino thiazine inhibitor LY2886721 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(4aS,7aS)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Timm, D.E. | | Deposit date: | 2014-12-09 | | Release date: | 2014-12-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Potent BACE1 Inhibitor LY2886721 Elicits Robust Central A beta Pharmacodynamic Responses in Mice, Dogs, and Humans.
J.Neurosci., 35, 2015
|
|
4YBI
 
 | | Crystal structure of BACE with amino thiazine inhibitor LY2811376 | | Descriptor: | (4S)-4-[2,4-difluoro-5-(pyrimidin-5-yl)phenyl]-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor.
J.Neurosci., 31, 2011
|
|
4ZSM
 
 | | BACE crystal structure with bicyclic aminothiazine fragment | | Descriptor: | (4aS,8aR)-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSQ
 
 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4S,4aS,6S,8aR)-10-aminohexahydro-3H-4,8a-(epithiomethenoazeno)isochromen-6(1H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSR
 
 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, N-[(4aS,6S,8aR)-2-amino-5,6,7,8-tetrahydro-4a,8a-(methanooxymethano)-3,1-benzothiazin-6(4H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSP
 
 | | BACE crystal structure with bicyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4aS,6S,8aR)-2-amino-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-6-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
1D1F
 
 | |
1D1E
 
 | |
1D0W
 
 | |
1QA5
 
 | |
1QA4
 
 | |
