1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | Descriptor: | Nucleoside Diphosphate Kinase A | | Authors: | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | Deposit date: | 2001-09-10 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
1PCV
 
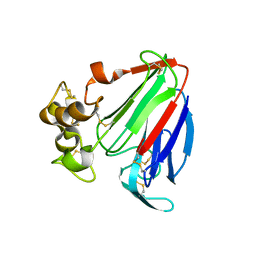 | | Crystal structure of osmotin, a plant antifungal protein | | Descriptor: | osmotin | | Authors: | Min, K, Ha, S.C, Yun, D.-J, Bressan, R.A, Kim, K.K. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of osmotin, a plant antifungal protein
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1EK8
 
 | |
7E60
 
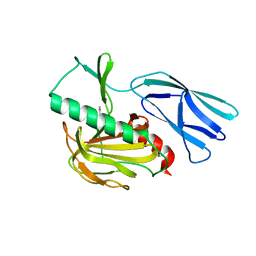 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | Descriptor: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
3CKP
 
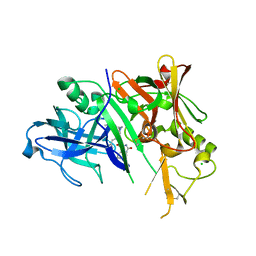 | | Crystal structure of BACE-1 in complex with inhibitor | | Descriptor: | (4S)-N-[(1S,2R)-1-benzyl-3-{[3-(dimethylamino)benzyl]amino}-2-hydroxypropyl]-1-(3-methoxybenzyl)-2-oxoimidazolidine-4-carboxamide, Beta-secretase 1, CHLORIDE ION | | Authors: | Min, K. | | Deposit date: | 2008-03-16 | | Release date: | 2008-06-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, SAR, and X-ray structure of human BACE-1 inhibitors with cyclic urea derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CKR
 
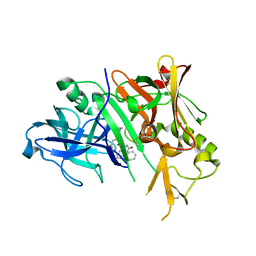 | | Crystal structure of BACE-1 in complex with inhibitor | | Descriptor: | (4S)-1,4-dibenzyl-N-[(1S,2R)-1-benzyl-3-{[3-(dimethylamino)benzyl]amino}-2-hydroxypropyl]-2-oxoimidazolidine-4-carboxamide, Beta-secretase 1 | | Authors: | Min, K. | | Deposit date: | 2008-03-16 | | Release date: | 2008-06-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, SAR, and X-ray structure of human BACE-1 inhibitors with cyclic urea derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3ENQ
 
 | |
3ENW
 
 | |
3ENV
 
 | |
4M47
 
 | | structure of human DNA polymerase complexed with 8-BrG in the template base paired with incoming non-hydrolyzable GTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4MF8
 
 | | Structure of human DNA polymerase beta complexed with nicked DNA containing a mismatched template O6MG and incoming CTP | | Descriptor: | DNA polymerase beta, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human DNA polymerase beta inserting bases opposite templating O6MG
To be Published
|
|
4M2Y
 
 | | Structure of human DNA polymerase beta complexed with 8-BrG as the template base in a 1-nucleotide gapped DNA | | Descriptor: | DNA polymerase beta, SODIUM ION, down-primer, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-05 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4MFA
 
 | | Structure of human DNA polymerase beta complexed with nicked DNA containing a mismatched template O6MG and incoming TTP | | Descriptor: | DNA polymerase beta, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of human DNA polymerase beta inserting bases opposite templating O6MG
To be Published
|
|
4NM1
 
 | | Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-C at N-1 position and G-C at N position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*(BGM)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4NM2
 
 | | Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-G at N-1 position and G-C at N position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*(BGM)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4NLK
 
 | | Structure of human DNA polymerase beta complexed with 8BrG in the template base-paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 5'-D(*CP*CP*GP*AP*CP*(BGM)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4NLN
 
 | | Structure of human DNA polymerase beta complexed with nicked DNA containing a template 8BrG and incoming CTP | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(BGM)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4NLZ
 
 | | Structure of human DNA polymerase beta complexed with nicked DNA containing a mismatched template 8BrG and incoming GTP | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(BGM)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*AP*G)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
3L7U
 
 | | Crystal structure of human NM23-H1 | | Descriptor: | Nucleoside diphosphate kinase A, PHOSPHATE ION | | Authors: | Han, B.G, Min, K, Lee, B.I, Lee, S. | | Deposit date: | 2009-12-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined Structure of Human NM23-H1 from a Hexagonal Crystal
BULL.KOREAN CHEM.SOC., 31, 2010
|
|
5YU4
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU3
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU0
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU1
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
6JN8
 
 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
