4AQY
 
 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3S
 
 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3M
 
 | | Crystal structure of the 30S ribosome in complex with compound 1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B3T
 
 | | Crystal structure of the 30S ribosome in complex with compound 39 | | Descriptor: | (2S,3S,4R,5R,6R)-2-(aminomethyl)-5-azanyl-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-bis(azanyl)-2-[(2S,3R,4R,5S,6R)-3-azanyl-5-[(4-chlorophenyl)methoxy]-6-(hydroxymethyl)-4-oxidanyl-oxan-2-yl]oxy-6-oxidanyl-cyclohexyl]oxy-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-oxane-3,4-diol, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
1JZU
 
 | | Cell transformation by the myc oncogene activates expression of a lipocalin: analysis of the gene (Q83) and solution structure of its protein product | | Descriptor: | lipocalin Q83 | | Authors: | Hartl, M, Matt, T, Schueler, W, Siemeister, G, Kontaxis, G, Kloiber, K, Konrat, R, Bister, K. | | Deposit date: | 2001-09-17 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Cell Transformation by the v-myc Oncogene Abrogates c-Myc/Max-mediated Suppression of a
C/EBPbeta-dependent Lipocalin Gene.
J.Mol.Biol., 333, 2003
|
|
1IBI
 
 | | QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, 15 MINIMIZED MODEL STRUCTURES | | Descriptor: | CYSTEINE-RICH PROTEIN 2, ZINC ION | | Authors: | Schuler, W, Kloiber, K, Matt, T, Bister, K, Konrat, R. | | Deposit date: | 2001-03-28 | | Release date: | 2001-09-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Application of cross-correlated NMR spin relaxation to the zinc-finger protein CRP2(LIM2): evidence for collective motions in LIM domains.
Biochemistry, 40, 2001
|
|
2HDP
 
 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
7R04
 
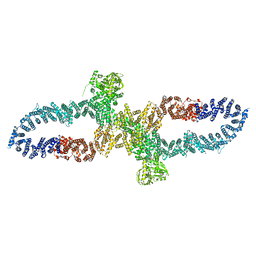 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
7R03
 
 | |
6EHR
 
 | |
6EHP
 
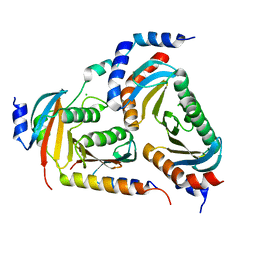 | | The crystal structure of the human LAMTOR complex | | Descriptor: | CHLORIDE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | Scheffzek, K, Naschberger, A. | | Deposit date: | 2017-09-14 | | Release date: | 2017-10-04 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling.
Science, 358, 2017
|
|
6FOG
 
 | |
6FOH
 
 | |
