1IZO
 
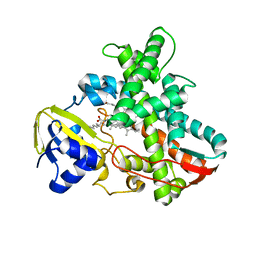 | | Cytochrome P450 BS beta Complexed with Fatty Acid | | Descriptor: | Cytochrome P450 152A1, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.S, Yamada, A, Sugimoto, H, Matsunaga, I, Ogura, H, Ichihara, K, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-10 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Molecular Mechanism of Fatty Acid Hydroxylation by Cytochrome P450 from Bacillus subtilis. CRYSTALLOGRAPHIC, SPECTROSCOPIC, AND MUTATIONAL STUDIES.
J.Biol.Chem., 278, 2003
|
|
2AEX
 
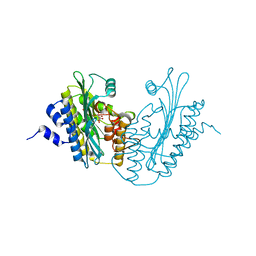 | | The 1.58A Crystal Structure of Human Coproporphyrinogen Oxidase Reveals the Structural Basis of Hereditary Coproporphyria | | Descriptor: | CITRIC ACID, Coproporphyrinogen III oxidase, mitochondrial | | Authors: | Lee, D.S, Flachsova, E, Bodnarova, M, Demeler, B, Martasek, P, Raman, C.S. | | Deposit date: | 2005-07-24 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of hereditary coproporphyria.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3DSK
 
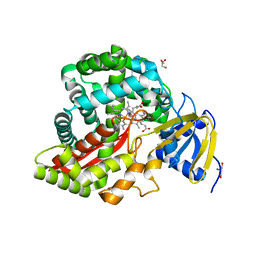 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase variant (F137L) (At-AOS(F137L), Cytochrome P450 74A, CYP74A) Complexed with 12R,13S-Vernolic Acid at 1.55 A Resolution | | Descriptor: | (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
3DSJ
 
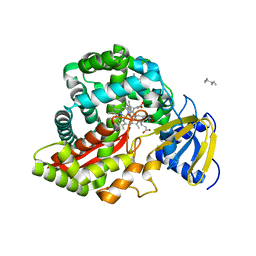 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase Variant (F137L) (At-AOS(F137L), cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOD at 1.60 A Resolution | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
3DSI
 
 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase (AOS, cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOT at 1.60 A resolution | | Descriptor: | (9Z,11E,13S,15Z)-13-hydroxyoctadeca-9,11,15-trienoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
2RCL
 
 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase (AOS, cytochrome P450 74A, CYP74A) Complexed with 12R,13S-Vernolic Acid at 2.4 A resolution | | Descriptor: | (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, Cytochrome P450 74A, GLYCEROL, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes.
Nature, 455, 2008
|
|
2RCM
 
 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase variant (F137L) (At-AOS(F137L), cytochrome P450 74A) at 1.73 A Resolution | | Descriptor: | Cytochrome P450 74A, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes.
Nature, 455, 2008
|
|
2RCH
 
 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase (AOS, cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOD at 1.85 A Resolution | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A, GLYCEROL, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes.
Nature, 455, 2008
|
|
8H24
 
 | | Leucine-rich alpha-2-glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich alpha-2-glycoprotein, SULFATE ION | | Authors: | Won, S.Y, Park, B.S, Lee, D.S, Kim, H.M, Han, A, Yang, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of LRG1 and the functional significance of LRG1 glycan for LPHN2 activation.
Exp.Mol.Med., 55, 2023
|
|
1IW0
 
 | | Crystal structure of a heme oxygenase (HmuO) from Corynebacterium diphtheriae complexed with heme in the ferric state | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Hirotsu, S, Unno, M, Chu, G.C, Lee, D.S, Park, S.Y, Shiro, Y, Ikeda-Saito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae.
J.Biol.Chem., 279, 2004
|
|
1IW1
 
 | | Crystal structure of a heme oxygenase (HmuO) from Corynebacterium diphtheriae complexed with heme in the ferrous state | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Hirotsu, S, Unno, M, Chu, G.C, Lee, D.S, Park, S.Y, Shiro, Y, Ikeda-Saito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-04 | | Release date: | 2003-04-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae.
J.Biol.Chem., 279, 2004
|
|
1V9Y
 
 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1V9Z
 
 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (Ferrous Form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1GEM
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450CAM, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
1GEK
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450CAM, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
1GEJ
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
1GEI
 
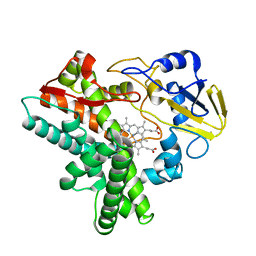 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-11-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
4Z8E
 
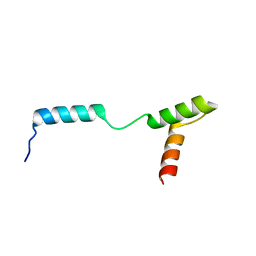 | | TEAD DBD mutant -deltaL1 | | Descriptor: | Transcriptional enhancer factor TEF-1 | | Authors: | Lee, D.-S, Albarado, D.C, Vonrhein, C, Raman, C.S, Veeraraghavan, S. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | A Potential Structural Switch for Regulating DNA-Binding by TEAD Transcription Factors.
J.Mol.Biol., 428, 2016
|
|
1BXB
 
 | | XYLOSE ISOMERASE FROM THERMUS THERMOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
1BXC
 
 | | XYLOSE ISOMERASE FROM THERMUS CALDOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
8H2G
 
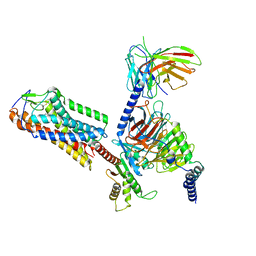 | |
8I7W
 
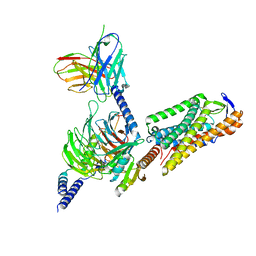 | | Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8I7V
 
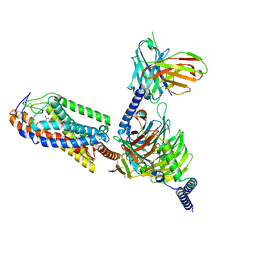 | | Cryo-EM structure of Acipimox bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8K5D
 
 | |
8K5B
 
 | |
