2QAP
 
 | |
2QDH
 
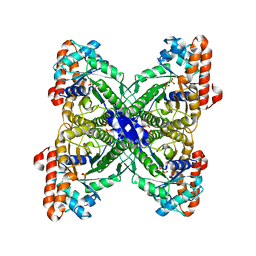
 | | Fructose-1,6-bisphosphate aldolase from Leishmania mexicana in complex with mannitol-1,6-bisphosphate, a competitive inhibitor | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, Fructose-1,6-bisphosphate aldolase | | Authors: | Lafrance-Vanasse, J, Sygusch, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carboxy-Terminus Recruitment Induced by Substrate Binding in Eukaryotic Fructose Bis-phosphate Aldolases
Biochemistry, 46, 2007
|
|
2QDG
 
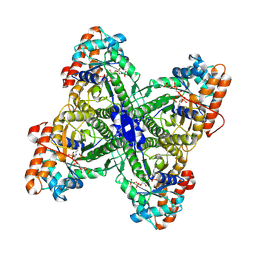 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from Leishmania mexicana | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-1,6-bisphosphate aldolase, PHOSPHATE ION | | Authors: | Lafrance-Vanasse, J, Sygusch, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carboxy-Terminus Recruitment Induced by Substrate Binding in Eukaryotic Fructose Bis-phosphate Aldolases
Biochemistry, 46, 2007
|
|
3F0P
 
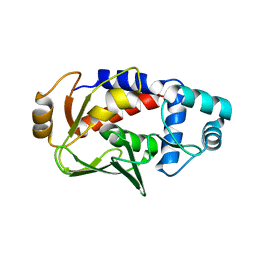 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F0O
 
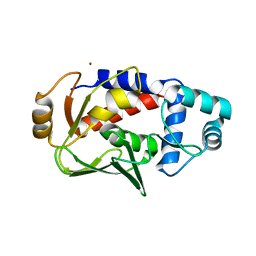 | | Crystal structure of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2H
 
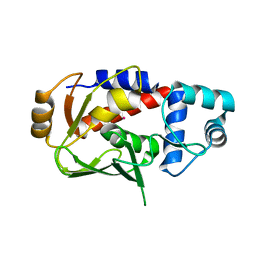 | | Crystal structure of the mercury-bound form of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2G
 
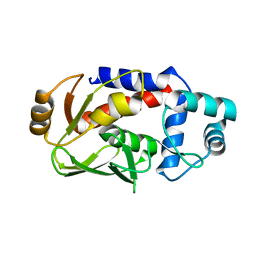 | | Crystal structure of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2F
 
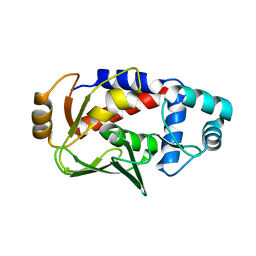 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
2LOX
 
 | |
2M14
 
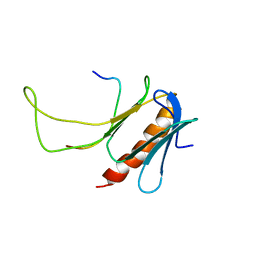 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and Rad4 | | Descriptor: | DNA repair protein RAD4, RNA polymerase II transcription factor B subunit 1 | | Authors: | Lafrance-Vanasse, J, Arseneault, G, Cappadocia, L, Legault, P, Omichinski, J.G. | | Deposit date: | 2012-11-16 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Rad4 competes with Rad2 for binding to the Tfb1 subunit of TFIIH in NER.
Nucleic Acids Res., 41, 2013
|
|
6MUJ
 
 | | Formylglycine generating enzyme bound to copper | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Lafrance-Vanasse, J, Appel, M.J, Tsai, C.-L, Bertozzi, C, Tainer, J.A. | | Deposit date: | 2018-10-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Formylglycine-generating enzyme binds substrate directly at a mononuclear Cu(I) center to initiate O2activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5DSF
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C17
 
 | | Crystal structure of the mercury-bound form of MerB2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-13 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0U
 
 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0T
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
1ZAI
 
 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from rabbit muscle | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZAJ
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with mannitol-1,6-bisphosphate, a competitive inhibitor | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZAH
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZAL
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, PHOSPHATE ION | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
2L2I
 
 | |
6N9T
 
 | | Structure of a peptide-based photo-affinity cross-linker with Herceptin Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin G1 FC, ... | | Authors: | Sadowsky, J, Ultsch, M, Vance, N, Wang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | Development, Optimization, and Structural Characterization of an Efficient Peptide-Based Photoaffinity Cross-Linking Reaction for Generation of Homogeneous Conjugates from Wild-Type Antibodies.
Bioconjug. Chem., 30, 2019
|
|
2MBH
 
 | | NMR structure of EKLF(22-40)/Ubiquitin Complex | | Descriptor: | Krueppel-like factor 1, Ubiquitin | | Authors: | Raiola, L, Omichinski, J.G. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a Noncovalent Complex between Ubiquitin and the Transactivation Domain of the Erythroid-Specific Factor EKLF.
Structure, 21, 2013
|
|
