3FRP
 
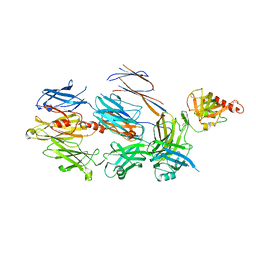 | | Crystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBb | | Descriptor: | CALCIUM ION, Cobra venom factor alpha chain, Cobra venom factor beta chain, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Crystal Structure of Cobra Venom Factor, a Cofactor for C3- and C5-Convertase CVFBb.
Structure, 17, 2009
|
|
3TVY
 
 | |
3TXA
 
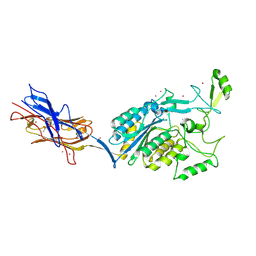 | | Structural Analysis of Adhesive Tip pilin, GBS104 from Group B Streptococcus agalactiae | | Descriptor: | CADMIUM ION, Cell wall surface anchor family protein, LITHIUM ION, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2011-09-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TW0
 
 | |
3ERB
 
 | |
3PHS
 
 | |
4JCN
 
 | |
2ODQ
 
 | | Complement component C2a, the catalytic fragment of C3- and C5-convertase of human complement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Narayana, S.V.L, Krishnan, V. | | Deposit date: | 2006-12-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of c2a, the catalytic fragment of classical pathway c3 and c5 convertase of human complement.
J.Mol.Biol., 367, 2007
|
|
2ODP
 
 | | Complement component C2a, the catalytic fragment of C3- and C5-convertase of human complement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Narayana, S.V.L, Krishnan, V. | | Deposit date: | 2006-12-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of c2a, the catalytic fragment of classical pathway c3 and c5 convertase of human complement.
J.Mol.Biol., 367, 2007
|
|
8GMX
 
 | |
7VCR
 
 | |
8GR6
 
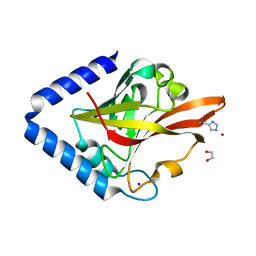 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
7W6B
 
 | |
7W7I
 
 | |
5HBB
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E139A mutant | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA, SODIUM ION, ... | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-31 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5HTS
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - D295N mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5J4M
 
 | | Crystal structure of shaft pilin SpaA from Lactobacillus rhamnosus GG - E269A/D295N double mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
7F7Y
 
 | |
7F87
 
 | |
7VCN
 
 | |
6JCH
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Orthorhombic form | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-28 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.536 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JBV
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Selenium derivative | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-26 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JK7
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Trigonal form | | Descriptor: | Pilus assembly protein | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6M48
 
 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - P21212 form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SpaC | | Authors: | Kant, A, Palva, A, Von Ossowaski, I, Krishnan, V. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6M3Y
 
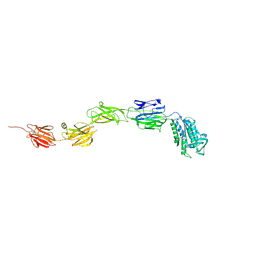 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - open conformation | | Descriptor: | MAGNESIUM ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
