4Z8L
 
 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | Descriptor: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | Authors: | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2015-04-09 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
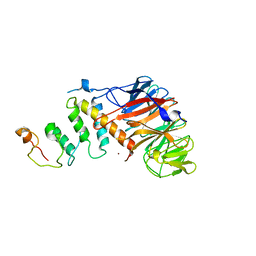
4JGF
 
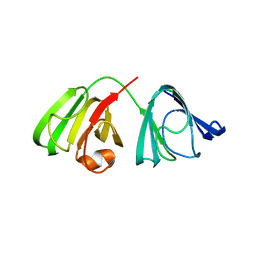 | |
3S5X
 
 | |
3S5V
 
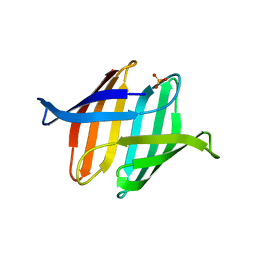 | |
3S60
 
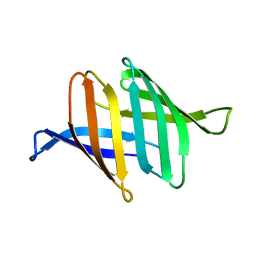 | |
4FBV
 
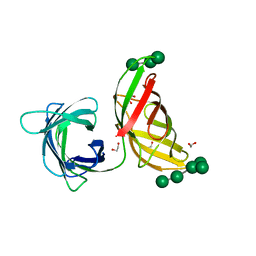 | | Crystal structure of the Myxococcus Xanthus hemagglutinin in complex with a3,a6-mannopentaose | | Descriptor: | 1,2-ETHANEDIOL, Myxobacterial hemagglutinin, alpha-D-mannopyranose, ... | | Authors: | Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into the Anti-HIV Activity of the Oscillatoria agardhii Agglutinin Homolog Lectin Family.
J.Biol.Chem., 287, 2012
|
|
4FBR
 
 | |
4FBO
 
 | |
3HNU
 
 | |
3OBL
 
 | |
3HNX
 
 | |
3HP8
 
 | | Crystal structure of a designed Cyanovirin-N homolog lectin; LKAMG, bound to sucrose | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cyanovirin-N-like protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Koharudin, L.M.I, Furey, W, Gronenborn, A.M. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A designed chimeric cyanovirin-N homolog lectin: Structure and molecular basis of sucrose binding.
Proteins, 77, 2009
|
|
5C8P
 
 | |
5C8O
 
 | |
5C8Q
 
 | | Crystal structure of MoCVNH3 variant (Mo0v) in complex with (N-GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MoCVNH3 variant | | Authors: | Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Fungal Cell Wall Recognition by a CVNH Protein with a Single LysM Domain.
Structure, 23, 2015
|
|
2JZJ
 
 | | Structure of CrCVNH (C. richardii CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZL
 
 | | Structure of NcCVNH (N. CRASSA CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZK
 
 | | Structure of TbCVNH (T. BORCHII CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2KJL
 
 | |
2L9Y
 
 | | Solution structure of the MoCVNH-LysM module from the rice blast fungus Magnaporthe oryzae protein (MGG_03307) | | Descriptor: | CVNH-LysM lectin | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Montanini, B, Kershaw, M.J, Talbot, N.J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2011-02-26 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Analysis of a CVNH-LysM Lectin Expressed during Plant Infection by the Rice Blast Fungus Magnaporthe oryzae.
Structure, 19, 2011
|
|
4J4E
 
 | |
4J4F
 
 | |
4J4G
 
 | |
4J4D
 
 | |
