7TCL
 
 | | Crystal structure of P.IsnB complexed with tyrosine isonitrile | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-2-isocyanopropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-12-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Elucidation of divergent desaturation pathways in the formation of vinyl isonitrile and isocyanoacrylate.
Nat Commun, 13, 2022
|
|
7N54
 
 | | Complex structure of GLAU4 with glaucine | | Descriptor: | GLAU4, SULFATE ION, glaucine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N4W
 
 | | Complex structure of ROTU4 with rotundine | | Descriptor: | GLYCEROL, ROTU4, SULFATE ION, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N4Z
 
 | | Complex structure of NOS4 with noscapine | | Descriptor: | NOS4, SULFATE ION, noscapine | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
7N53
 
 | | Complex structure of PAP4 with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, GLYCEROL, PAP4, ... | | Authors: | Kim, W, Zhang, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fungible biosensors to evolve improved alkaloid biosyntheses.
Nat.Chem.Biol., 18, 2022
|
|
8UKE
 
 | |
7SH6
 
 | |
8CU6
 
 | | Crystal structure of A2AAR-StaR2-S277-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
8CU7
 
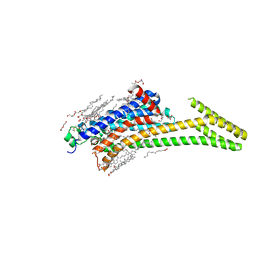 | | Crystal structure of A2AAR-StaR2-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
1JI7
 
 | | Crystal Structure of TEL SAM Polymer | | Descriptor: | ETS-RELATED PROTEIN TEL1, SULFATE ION | | Authors: | Kim, C.A, Phillips, M.L, Kim, W, Gingery, M, Tran, H.H, Robinson, M.A, Faham, S, Bowie, J.U. | | Deposit date: | 2001-06-29 | | Release date: | 2002-07-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression.
EMBO J., 20, 2001
|
|
1PK1
 
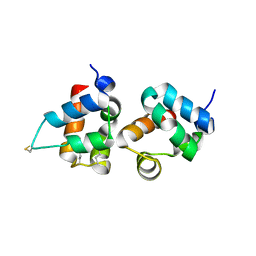 | | Hetero SAM domain structure of Ph and Scm. | | Descriptor: | Polyhomeotic-proximal chromatin protein, Sex comb on midleg CG9495-PA | | Authors: | Kim, C.A, Sawaya, M.R, Cascio, D, Kim, W, Bowie, J.U. | | Deposit date: | 2003-06-04 | | Release date: | 2005-02-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer.
J.Biol.Chem., 280, 2005
|
|
1PK3
 
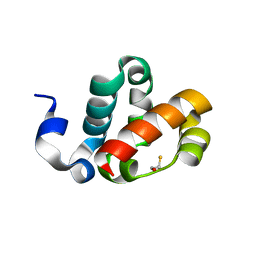 | | Scm SAM domain | | Descriptor: | BETA-MERCAPTOETHANOL, Sex comb on midleg CG9495-PA | | Authors: | Kim, C.A, Sawaya, M.R, Cascio, D, Kim, W, Bowie, J.U. | | Deposit date: | 2003-06-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer.
J.Biol.Chem., 280, 2005
|
|
8U8Q
 
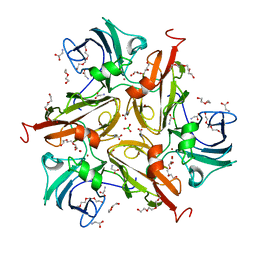 | | V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8P
 
 | | S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8R
 
 | | Y229F/V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8T
 
 | | Y229F/V290N Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8S
 
 | | Y229F/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QDN
 
 | |
8EZ7
 
 | | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 1F04, Light chain of influenza virus neuraminidase antibody 1F04, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase
Immunity, 56, 2023
|
|
8EZ3
 
 | | Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3A10, Light chain of influenza virus neuraminidase antibody 3A10, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase
Immunity, 56, 2023
|
|
8EZ8
 
 | | Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3C08, Light chain of influenza virus neuraminidase antibody 3C08, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase
Immunity, 56, 2023
|
|
7QZU
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 47 | | Descriptor: | (2~{S})-2-[2-[4-[3,4-bis(oxidanyl)-9,10-bis(oxidanylidene)anthracen-2-yl]sulfonylpiperazin-1-yl]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2022-01-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
1EK8
 
 | |
