5K60
 
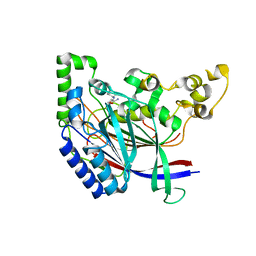 | | Crystal structure of N-terminal amidase with Gln-Val peptide | | Descriptor: | GLUTAMINE, Nta1p, VALINE | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K66
 
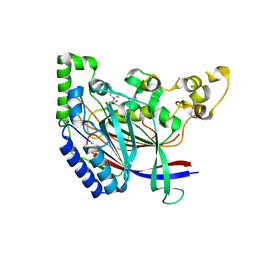 | | Crystal structure of N-terminal amidase with Asn-Glu peptide | | Descriptor: | ASPARAGINE, GLUTAMIC ACID, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K62
 
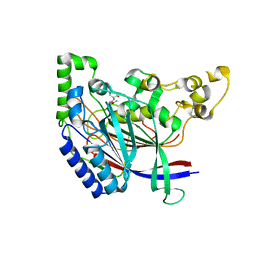 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, Nta1p, VALINE | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K5V
 
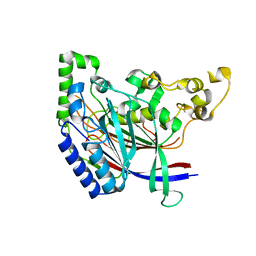 | |
5K61
 
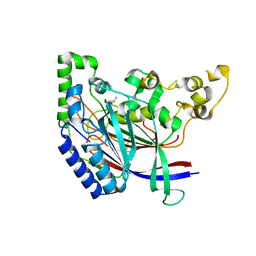 | | Crystal structure of N-terminal amidase with Gln-Gly peptide | | Descriptor: | GLUTAMINE, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2B7Q
 
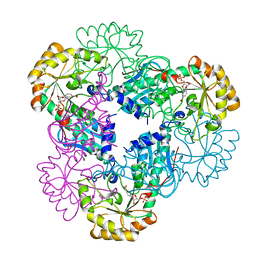 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with nicotinate mononucleotide | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Probable nicotinate-nucleotide pyrophosphorylase | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-05 | | Release date: | 2006-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B7P
 
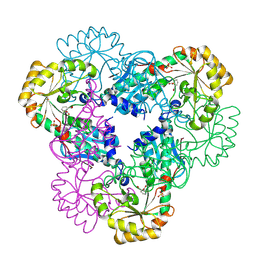 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with phthalic acid | | Descriptor: | PHTHALIC ACID, Probable nicotinate-nucleotide pyrophosphorylase, SULFATE ION | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-05 | | Release date: | 2006-02-14 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B7N
 
 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori | | Descriptor: | Probable nicotinate-nucleotide pyrophosphorylase, QUINOLINIC ACID, SULFATE ION | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-04 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
5HYY
 
 | |
5K5U
 
 | |
5K63
 
 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, GLYCINE, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5B62
 
 | | Crystal structure of N-terminal amidase with Asn-Glu-Ala peptide | | Descriptor: | ASN-GLU-ALA, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4J4K
 
 | | Crystal structure of glucose isomerase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Xylose isomerase, ... | | Authors: | Kim, M.K, An, Y.J, Lee, S, Jeong, C.S, Cha, S.S. | | Deposit date: | 2013-02-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glucose isomerase
To be Published
|
|
7WG4
 
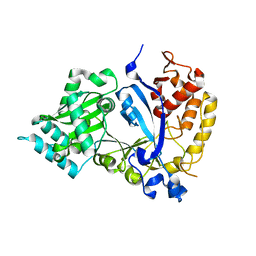 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WFX
 
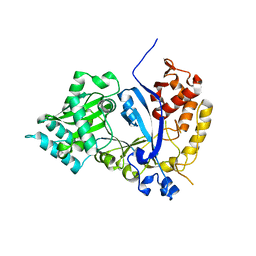 | | EVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-27 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG1
 
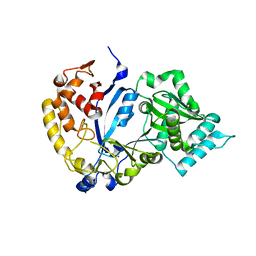 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-27 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG2
 
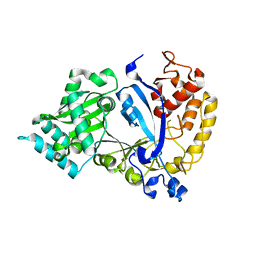 | | EVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5F1G
 
 | | Crystal structure of AmpC BER adenylylated in the cytoplasm | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Beta-lactamase, ... | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5F1F
 
 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
7TM3
 
 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TUT
 
 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P81
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
2ZCU
 
 | | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from escherichia coli | | Descriptor: | COPPER (II) ION, Uncharacterized oxidoreductase ytfG | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
2ZCV
 
 | | Crystal structure of NADPH-dependent quinone oxidoreductase QOR2 complexed with NADPH from escherichia coli | | Descriptor: | COPPER (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
