3MYR
 
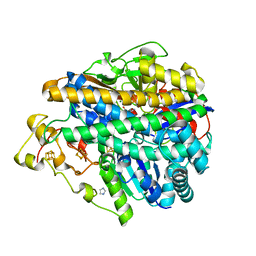 | | Crystal structure of [NiFe] hydrogenase from Allochromatium vinosum in its Ni-A state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Hydrogenase (NiFe) small subunit HydA, ... | | Authors: | Ogata, H, Kellers, P, Lubitz, W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni-A state).
J.Mol.Biol., 402, 2010
|
|
4ASL
 
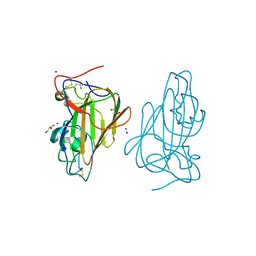 | | Structure of Epa1A in complex with the T-antigen (Gal-b1-3- GalNAc) | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V56
 
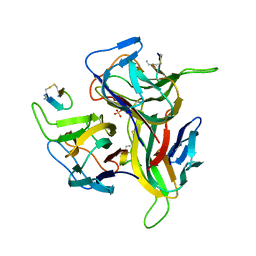 | | Re-refinement of PDB entry 1OSG - Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold - reveals an additonal copy of the peptide. | | Descriptor: | BR3 derived peptive, SULFATE ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Smart, O.S, Womack, T.O, Flensburg, C, Keller, P, Sharff, A, Paciorek, W, Vonrhein, C, Bricogne, G. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3URP
 
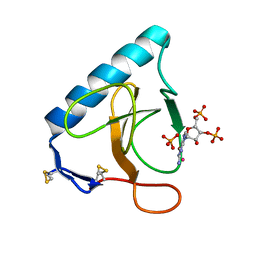 | | Re-refinement of PDB entry 5RNT - ribonuclease T1 with guanosine-3',5'-diphosphate and phosphate ion bound | | Descriptor: | GUANOSINE-3',5'-DIPHOSPHATE, Guanyl-specific ribonuclease T1, PHOSPHATE ION, ... | | Authors: | Smart, O.S, Womack, T.O, Flensburg, C, Keller, P, Sharff, A, Paciorek, W, Vonrhein, C, Bricogne, G. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AFC
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 6 A domain (Epa1to6A) from Candida glabrata in complex with Galb1-3Glc | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, EPA1P, ... | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFB
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 3 A domain (Epa1to3A) from Candida glabrata in complex with glycerol | | Descriptor: | CALCIUM ION, EPA1P, GLYCEROL | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AF9
 
 | | Crystal Structure of Epithelial Adhesin 1 A domain (Epa1A) from Candida glabrata in complex with Galb1-3Glc | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, EPA1P, ... | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFA
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 2 A domain (Epa1to2A) from Candida glabrata in complex with glycerol | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, EPA1P, ... | | Authors: | Maestre-Reyna, M, Diderrich, R, Veelders, M.S, Eulenburg, G, Kalugin, V, Brueckner, S, Keller, P, Rupp, S, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Promiscuity and Specificity During Candida Glabrata Invasion of Host Epithelia.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5V3Z
 
 | |
5V41
 
 | |
5V42
 
 | |
5V3Y
 
 | |
5V3W
 
 | |
5V3X
 
 | |
5V40
 
 | |
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
7ZFM
 
 | | Engineered Protein Targeting the Zika Viral Envelope Fusion Loop | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Athayde, D, Archer, M, Viana, I.F.T, Adan, W.C.S, Xavier, L.S.S, Lins, R.D. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | In Vitro Neutralisation of Zika Virus by an Engineered Protein Targeting the Viral Envelope Fusion Loop
SSRN, 2022
|
|
3SYU
 
 | | Re-refined coordinates for pdb entry 1det - ribonuclease T1 carboxymethylated at GLU 58 in complex with 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1, SODIUM ION, ... | | Authors: | Smart, O.S, Womack, T.O, Bricogne, G. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8SKQ
 
 | |
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
8A67
 
 | |
6RTD
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C, ... | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
