1ETJ
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLU | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Karlsson, B.G, Tsai, L.-C, Nar, H, Sanders-Loehr, J, Bonander, N, Langer, V, Sjolin, L. | | Deposit date: | 1997-01-11 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure determination and characterization of the Pseudomonas aeruginosa azurin mutant Met121Glu.
Biochemistry, 36, 1997
|
|
8S03
 
 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Method: | SOLUTION NMR | | Cite: | The second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking
To Be Published
|
|
6TPB
 
 | | NMR structure of the apo-form of Pseudomonas fluorescens CopC | | Descriptor: | Putative copper resistance protein | | Authors: | Persson, K.C, Mayzel, M, Karlsson, B.G, Peciulyte, A, Olsson, L, Wittung Stafshede, P, Salomon Johansen, K, Horvath, I. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Pseudomonas fluorescens CopC
To Be Published
|
|
2M7L
 
 | |
2PCF
 
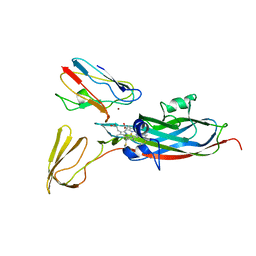 | | THE COMPLEX OF CYTOCHROME F AND PLASTOCYANIN DETERMINED WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | COPPER (II) ION, CYTOCHROME F, HEME C, ... | | Authors: | Ubbink, M, Ejdeback, M, Karlsson, B.G, Bendall, D.S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics.
Structure, 6, 1998
|
|
4CSQ
 
 | |
2XNA
 
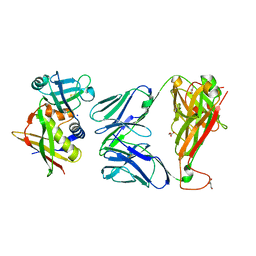 | | Crystal structure of the complex between human T cell receptor and staphylococcal enterotoxin | | Descriptor: | ENTEROTOXIN H, GLYCEROL, SODIUM ION, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
2XN9
 
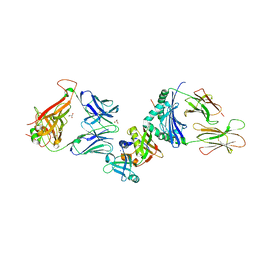 | | Crystal structure of the ternary complex between human T cell receptor, staphylococcal enterotoxin H and human major histocompatibility complex class II | | Descriptor: | ENTEROTOXIN H, GLYCEROL, HEMAGGLUTININ, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
1X13
 
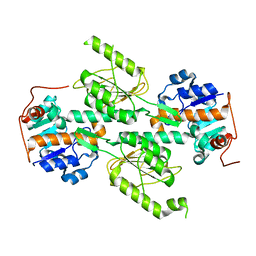 | | Crystal structure of E. coli transhydrogenase domain I | | Descriptor: | NAD(P) transhydrogenase subunit alpha | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1X14
 
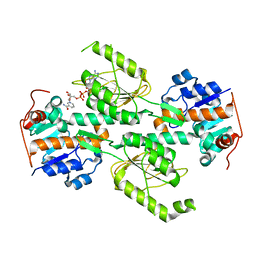 | | Crystal structure of E. coli transhydrogenase domain I with bound NAD | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1X15
 
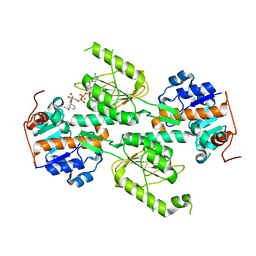 | | Crystal structure of E. coli transhydrogenase domain I with bound NADH | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1EZL
 
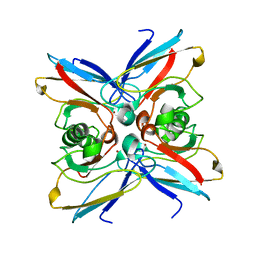 | | CRYSTAL STRUCTURE OF THE DISULPHIDE BOND-DEFICIENT AZURIN MUTANT C3A/C26A: HOW IMPORTANT IS THE S-S BOND FOR FOLDING AND STABILITY? | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Bonander, N, Leckner, J, Guo, H, Karlsson, B.G, Sjolin, L. | | Deposit date: | 2000-05-11 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the disulfide bond-deficient azurin mutant C3A/C26A: how important is the S-S bond for folding and stability?
Eur.J.Biochem., 267, 2000
|
|
1GR7
 
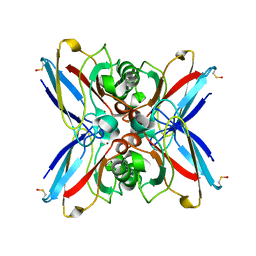 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
2BRU
 
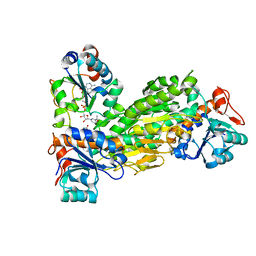 | | Complex of the domain I and domain III of Escherichia coli transhydrogenase | | Descriptor: | NAD(P) TRANSHYDROGENASE SUBUNIT ALPHA, NAD(P) TRANSHYDROGENASE SUBUNIT BETA, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Johansson, T, Pedersen, A, Leckner, J, Karlsson, B.G. | | Deposit date: | 2005-05-11 | | Release date: | 2006-08-29 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of a Transient Complex by NMR Using Paramagnetic Distance Restraints - the Complex of the Soluble Domains of Escherichia Coli Transhydrogenase
To be Published
|
|
1NZR
 
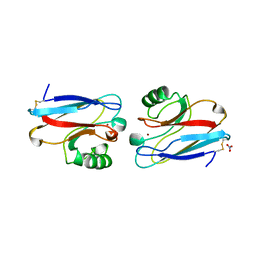 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
5ABK
 
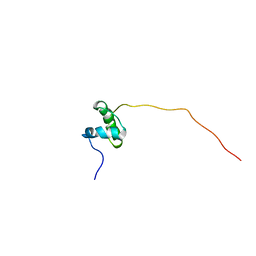 | | Structure of the N-terminal domain of the metalloprotease PrtV from Vibrio cholerae | | Descriptor: | METALLOPROTEASE | | Authors: | Persson, C, Mayzel, M, Edwin, A, Wai, S.N, Ohman, A, Sauer-Eriksson, A.E, Karlsson, G. | | Deposit date: | 2015-08-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal Domain of the Metalloprotease Prtv from Vibrio Cholerae.
Protein Sci., 24, 2015
|
|
8PH4
 
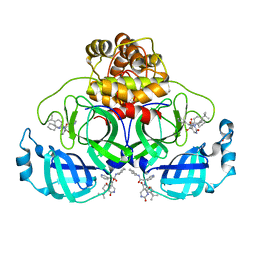 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
5LFI
 
 | | lactococcin A immunity protein | | Descriptor: | Lactococcin-A immunity protein | | Authors: | Persson, C, Fuochi, V, Pedersen, A, Karlsson, B.G, Nissen-Meyer, J, Kristiansen, P.E, Oppegard, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure and Mutational Analysis of the Lactococcin A Immunity Protein.
Biochemistry, 55, 2016
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2TSA
 
 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1VLX
 
 | | STRUCTURE OF ELECTRON TRANSFER (COBALT-PROTEIN) | | Descriptor: | AZURIN, COBALT (II) ION | | Authors: | Bonander, N, Vanngard, T, Tsai, L.-C, Langer, V, Nar, H, Sjolin, L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The metal site of Pseudomonas aeruginosa azurin, revealed by a crystal structure determination of the Co(II) derivative and Co-EPR spectroscopy.
Proteins, 27, 1997
|
|
