3JSK
 
 | | Thiazole synthase from Neurospora crassa | | Descriptor: | ADENOSINE DIPHOSPHATE 5-(BETA-ETHYL)-4-METHYL-THIAZOLE-2-CARBOXYLIC ACID, CyPBP37 protein, FE (II) ION | | Authors: | Kang, Y.N, Bale, S, Ealick, S.E. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of thiazole synthase Thi4 from Neurospora crassa
to be published
|
|
3KHS
 
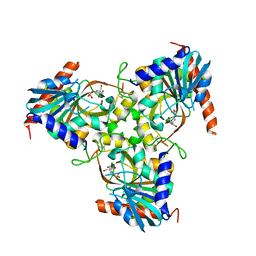 | | Crystal structure of grouper iridovirus purine nucleoside phosphorylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Kang, Y.N, Zhang, Y, Allan, P.W, Parker, W.B, Ting, J.W, Chang, C.Y, Ealick, S.E. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of grouper iridovirus purine nucleoside phosphorylase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2NTM
 
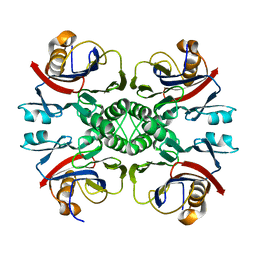 | |
2NTK
 
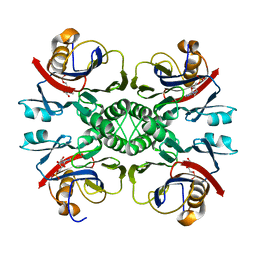 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2NTL
 
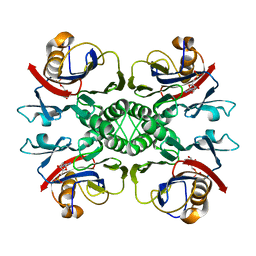 | | Crystal structure of PurO/AICAR from Methanothermobacter thermoautotrophicus | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, IMP cyclohydrolase | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
1UKP
 
 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-31 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1UKO
 
 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-30 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1V3H
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1V3I
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1WDS
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDP
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDR
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDQ
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
3SW4
 
 | |
3SW7
 
 | |
4YKQ
 
 | | Heat Shock Protein 90 Bound to CS301 | | Descriptor: | 1-(5-ethyl-2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS301
To Be Published
|
|
4YKY
 
 | | Heat Shock Protein 90 Bound to CS319 | | Descriptor: | (2,4-dihydroxyphenyl)(4-hydroxyphenyl)methanone, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS319
To Be Published
|
|
4YKZ
 
 | | Heat Shock Protein 90 Bound to CS320 | | Descriptor: | (2,4-dihydroxyphenyl)(3-hydroxyphenyl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS320
To Be Published
|
|
4YKT
 
 | | Heat Shock Protein 90 Bound to CS307 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-5-({[dihydroxy(pyridin-3-yl)-lambda~4~-sulfanyl]amino}methyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS307
To Be Published
|
|
4YKW
 
 | | Heat Shock Protein 90 Bound to CS312 | | Descriptor: | 4-(2-chloro-4-nitrophenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS312
To Be Published
|
|
4YKR
 
 | | Heat Shock Protein 90 Bound to CS302 | | Descriptor: | 1-(2,4-dihydroxy-5-propylphenyl)-1,3-dihydro-2H-benzimidazol-2-one, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS302
To Be Published
|
|
4YKX
 
 | | Heat Shock Protein 90 Bound to CS318 | | Descriptor: | (5-chloro-2-hydroxyphenyl)(4-hydroxyphenyl)methanone, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS318
To Be Published
|
|
4YKU
 
 | | Heat Shock Protein 90 Bound to CS311 | | Descriptor: | 6-(2-chlorophenyl)-1,3,5-triazine-2,4-diamine, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS311
To Be Published
|
|
4EK5
 
 | |
4EK8
 
 | |
