7COE
 
 | | Crystal structure of Receptor binding domain of MERS-CoV and KNIH90-F1 Fab complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Lee, J.Y, Song, J.Y, Lee, H.S, Hong, E, Jang, T.H. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a novel antibody against the spike protein inhibits Middle East respiratory syndrome coronavirus infections.
Sci Rep, 12, 2022
|
|
4OM7
 
 | | Crystal structure of TIR domain of TLR6 | | Descriptor: | Toll-like receptor 6 | | Authors: | Park, H.H, Jang, T.H. | | Deposit date: | 2014-01-26 | | Release date: | 2014-08-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal Structure of TIR Domain of TLR6 Reveals Novel Dimeric Interface of TIR-TIR Interaction for Toll-Like Receptor Signaling Pathway.
J.Mol.Biol., 426, 2014
|
|
4PYG
 
 | | Transglutaminase2 complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Park, H.H, Jang, T.H. | | Deposit date: | 2014-03-27 | | Release date: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of transglutaminase 2 with GTP complex and amino acid sequence evidence of evolution of GTP binding site.
Plos One, 9, 2014
|
|
7CM0
 
 | | Crystal structure of a glutaminyl cyclase in complex with NHV-1009 | | Descriptor: | 1-(cyclopentylmethyl)-1-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]-3-[3-(5-methylimidazol-1-yl)propyl]urea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Lee, J.W, Song, J.Y, Jang, T.H, Ha, J.H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of highly potent human glutaminyl cyclase (QC) inhibitors as anti-Alzheimer's agents by the combination of pharmacophore-based and structure-based design.
Eur.J.Med.Chem., 226, 2021
|
|
6ILQ
 
 | | Crystal structure of PPARgamma with compound BR101549 | | Descriptor: | Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma, ethyl [2-butyl-6-oxo-1-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}-4-(propan-2-yl)-1,6-dihydropyrimidin-5-yl]acetate | | Authors: | Hong, E, Jang, T.H, Chin, J, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-10-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Identification of BR101549 as a lead candidate of non-TZD PPAR gamma agonist for the treatment of type 2 diabetes: Proof-of-concept evaluation and SAR.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4JUP
 
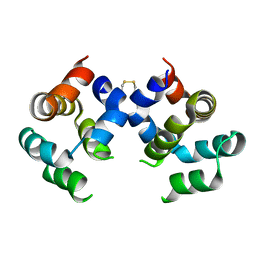 | | Dimeric structure of CARMA1 CARD | | Descriptor: | Caspase recruitment domain-containing protein 11 | | Authors: | Park, H.H. | | Deposit date: | 2013-03-25 | | Release date: | 2014-02-05 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel Disulfide Bond-Mediated Dimerization of the CARD Domain Was Revealed by the Crystal Structure of CARMA1 CARD
Plos One, 8, 2013
|
|
4D2K
 
 | |
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | Descriptor: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | Authors: | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | Deposit date: | 2014-09-04 | | Release date: | 2015-07-01 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
5ZOO
 
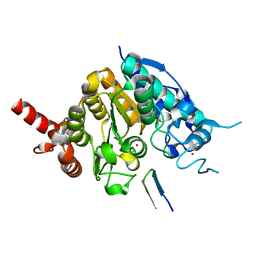 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP1 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP1 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
5ZOP
 
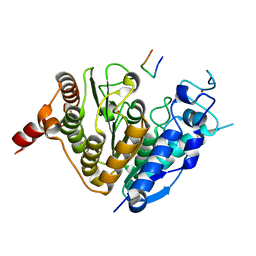 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP2 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP2 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
4MAC
 
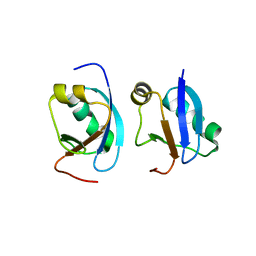 | | Crystal structure of CIDE-N domain of FSP27 | | Descriptor: | Cell death activator CIDE-3 | | Authors: | Park, H.H, Lee, S.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for homo-dimerization of the CIDE domain revealed by the crystal structure of the CIDE-N domain of FSP27
Biochem.Biophys.Res.Commun., 439, 2013
|
|
5ZYN
 
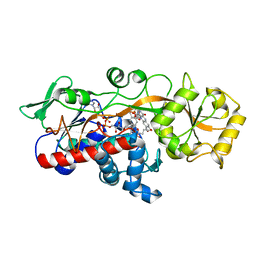 | | Fumarate reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fumarate reductase 2, ... | | Authors: | Park, H.H, Kim, C.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of maintaining an oxidizing environment under anaerobiosis by soluble fumarate reductase.
Nat Commun, 9, 2018
|
|
5GLG
 
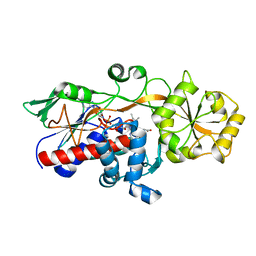 | |
6ICJ
 
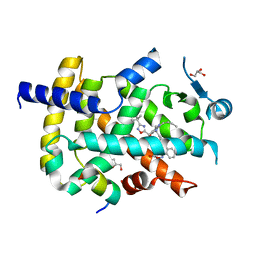 | | Crystal structure of PPARgamma with compound BR102375K | | Descriptor: | 2-butyl-5-[(3-tert-butyl-1,2,4-oxadiazol-5-yl)methyl]-6-methyl-3-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}pyrimidin-4(3H)-one, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Hong, E, Chin, J, Jang, T.H, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structure of PPARgamma with compound BR102375K
To Be Published
|
|
