4TRD
 
 | |
1D4L
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1D4K
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
1D31
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF BULGE-CONTAINING DNA FRAGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Joshua-Tor, L, Frolow, F, Appella, E, Hope, H, Rabinovich, D, Sussman, J.L. | | Deposit date: | 1991-04-25 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of bulge-containing DNA fragments.
J.Mol.Biol., 225, 1992
|
|
4TPL
 
 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OXTOXYNOL-10, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2014-06-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2AAS
 
 | | HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE A IN SOLUTION BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | RIBONUCLEASE A | | Authors: | Santoro, J, Gonzalez, C, Bruix, M, Neira, J.L, Nieto, J.L, Herranz, J, Rico, M. | | Deposit date: | 1992-11-20 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 229, 1993
|
|
4UBE
 
 | | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C. | | Deposit date: | 2014-08-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | CRYSTAL STRUCTURE OF M TUBERCULOSIS ADENOSINE KINASE COMPLEXED WITH 2-FLURO ADENOSINE
TO BE PUBLISHED
|
|
6XSG
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66T at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B, Sorenson, J.L. | | Deposit date: | 2020-07-15 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66T at cryogenic temperature
To be Published
|
|
2WWD
 
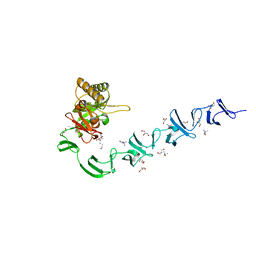 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WW5
 
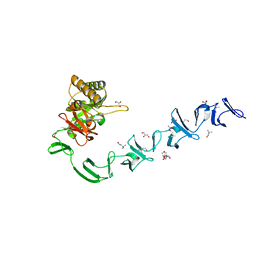 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WWC
 
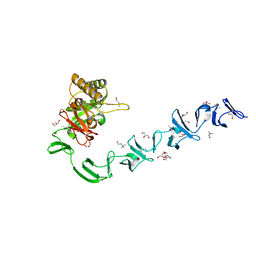 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHOLINE ION, GLYCEROL | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1CI3
 
 | | CYTOCHROME F FROM THE B6F COMPLEX OF PHORMIDIUM LAMINOSUM | | Descriptor: | HEME C, PROTEIN (CYTOCHROME F), ZINC ION | | Authors: | Carrell, C.J, Schlarb, B.G, Howe, C.J, Bendall, D.S, Cramer, W.A, Smith, J.L. | | Deposit date: | 1999-04-07 | | Release date: | 1999-08-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum.
Biochemistry, 38, 1999
|
|
1CFJ
 
 | | METHYLPHOSPHONYLATED ACETYLCHOLINESTERASE (AGED) OBTAINED BY REACTION WITH O-ISOPROPYLMETHYLPHOSPHONOFLUORIDATE (GB, SARIN) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METHYLPHOSPHONIC ACID ESTER GROUP, PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Millard, C.B, Silman, I, Sussman, J.L. | | Deposit date: | 1999-03-19 | | Release date: | 1999-06-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level.
Biochemistry, 38, 1999
|
|
1CTM
 
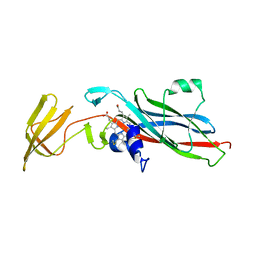 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
1A2M
 
 | |
1BOQ
 
 | | PRO REGION C-TERMINUS: PROTEASE ACTIVE SITE INTERACTIONS ARE CRITICAL IN CATALYZING THE FOLDING OF ALPHA-LYTIC PROTEASE | | Descriptor: | PROTEIN (ALPHA-LYTIC PROTEASE), SULFATE ION | | Authors: | Peters, R.J, Shiau, A.K, Sohl, J.L, Anderson, D.E, Tang, G, Silen, J.L, Agard, D.A. | | Deposit date: | 1998-08-05 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pro region C-terminus:protease active site interactions are critical in catalyzing the folding of alpha-lytic protease.
Biochemistry, 37, 1998
|
|
1HSB
 
 | | DIFFERENT LENGTH PEPTIDES BIND TO HLA-AW68 SIMILARLY AT THEIR ENDS BUT BULGE OUT IN THE MIDDLE | | Descriptor: | ALANINE, ARGININE, BETA 2-MICROGLOBULIN, ... | | Authors: | Guo, H.-C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle.
Nature, 360, 1992
|
|
1J7C
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E95K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1A2J
 
 | | OXIDIZED DSBA CRYSTAL FORM II | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1J7A
 
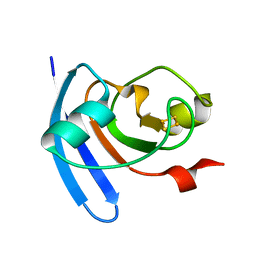 | | STRUCTURE OF THE ANABAENA FERREDOXIN D68K MUTANT | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, VanHooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollen, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1J7B
 
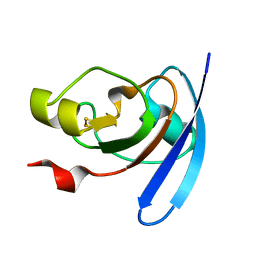 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E94K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1A2L
 
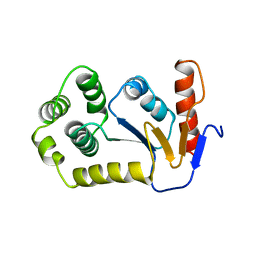 | | REDUCED DSBA AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
5I9O
 
 | |
5K5P
 
 | |
4R3V
 
 | | Structure of karilysin propeptide and catalytic MMP domain | | Descriptor: | CALCIUM ION, GLYCEROL, Karilysin, ... | | Authors: | Lopez-Pelegrin, M, Ksiazek, M, Karim, A.Y, Guevara, T, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2014-08-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel mechanism of latency in matrix metalloproteinases.
J.Biol.Chem., 290, 2015
|
|
