6TWT
 
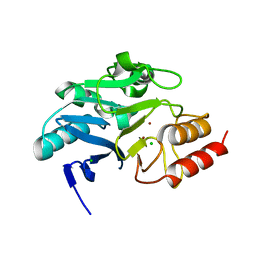 | | Crystal structure of N-terminally truncated NDM-1 metallo-beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Imiolczyk, B, Czyrko-Horczak, J, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
7OU1
 
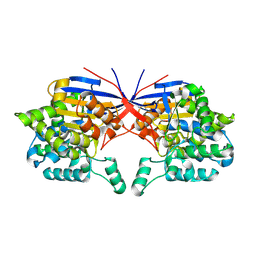 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV (monoclinic form MP2) | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, ZINC ION | | Authors: | Imiolczyk, B, Loch, J.I, Gilski, M, Jaskolski, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
6OL8
 
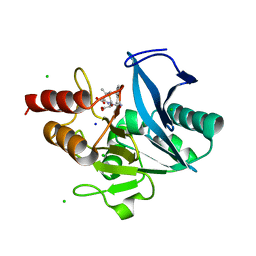 | | Crystal structure of NDM-12 metallo-beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-12, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
6OGO
 
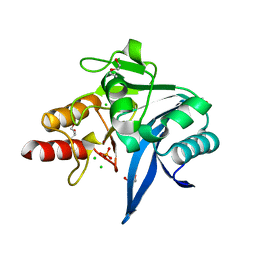 | | Crystal structure of NDM-9 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
8OSW
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (R4mC-1) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8ORI
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAIV (orthorhombic) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CLZ
 
 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (monoclinic form R4mC-2) | | Descriptor: | CHLORIDE ION, Putative L-asparaginase II protein, ZINC ION | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CLY
 
 | | Crystal structure of Rhizobium etli constitutive L-asparaginase ReAIV (tetragonal form R4tP) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative L-asparaginase II protein, ... | | Authors: | Loch, J.I, Worsztynowicz, P, Sliwiak, J, Imiolczyk, B, Grzechowiak, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7ZD8
 
 | | Crystal structure of the R24E mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD7
 
 | | Crystal structure of the R24E/E352T double mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD9
 
 | | Crystal structure of the E352T mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4PV3
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, SODIUM ION | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PU6
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ cations | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PV2
 
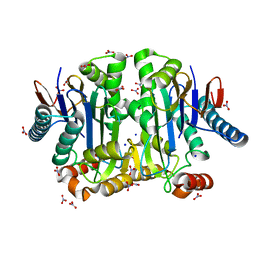 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7OS6
 
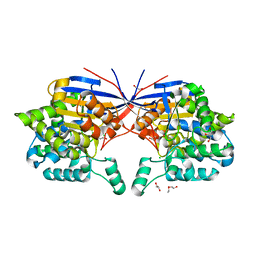 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV (monoclinic form MP1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Loch, J.I, Imiolczyk, B, Gilski, M, Jaskolski, M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-11-24 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
7OS3
 
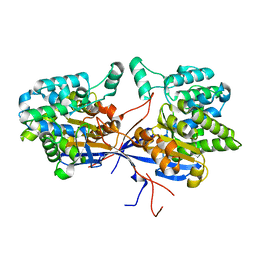 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV solved by S-SAD (orthorhombic form START) | | Descriptor: | CHLORIDE ION, L-asparaginase II protein, ZINC ION | | Authors: | Gilski, M, Loch, J.I, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
7OS5
 
 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV (orthorhombic form OP) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L-asparaginase, ... | | Authors: | Loch, J.I, Imiolczyk, B, Gilski, M, Jaskolski, M. | | Deposit date: | 2021-06-07 | | Release date: | 2021-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
7O5M
 
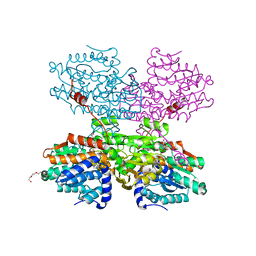 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Na+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7O5L
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7OZ6
 
 | | Crystal structure of Rhizobium etli inducible L-asparaginase ReAV (monoclinic form MC) | | Descriptor: | DI(HYDROXYETHYL)ETHER, L-asparaginase, ZINC ION | | Authors: | Gilski, M, Loch, J.I, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2021-06-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | Crystal structures of the elusive Rhizobium etli L-asparaginase reveal a peculiar active site.
Nat Commun, 12, 2021
|
|
7QY6
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYM
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-18 (R207V, D210P, S211W) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYX
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-24 (R207A, D210S, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R5C
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | Authors: | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
