1YZ0
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
1YXI
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-21 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
1YYZ
 
 | | R-State AMP Complex Reveals Initial Steps of the Quaternary Transition of Fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase, ... | | Authors: | Iancu, C.V, Mukund, S, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R-state AMP complex reveals initial steps of the quaternary transition of fructose-1,6-bisphosphatase.
J.Biol.Chem., 280, 2005
|
|
2DGN
 
 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
1IWE
 
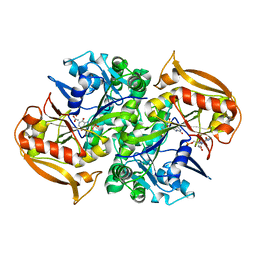 | | IMP Complex of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase | | Descriptor: | ACETATE ION, Adenylosuccinate Synthetase, INOSINIC ACID, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-05-04 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1J4B
 
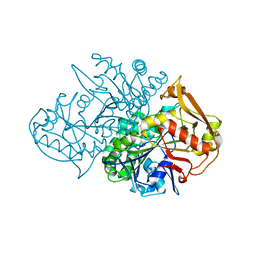 | | Recombinant Mouse-Muscle Adenylosuccinate Synthetase | | Descriptor: | adenylosuccinate synthetase | | Authors: | Iancu, C.V, Borza, T, Choe, J.Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-09-07 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recombinant mouse muscle adenylosuccinate synthetase: overexpression, kinetics, and crystal structure.
J.Biol.Chem., 276, 2001
|
|
1LNY
 
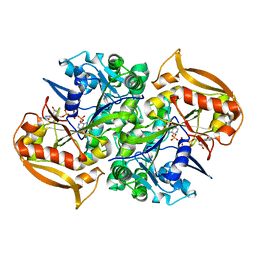 | | Crystal structure of the recombinant mouse-muscle adenylosuccinate synthetase complexed with 6-phosphoryl-IMP, GDP and Mg | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-05-04 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1LON
 
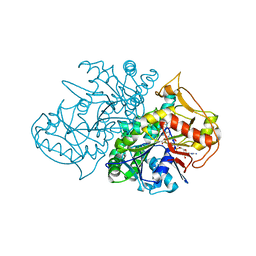 | | Crystal Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with 6-phosphoryl-IMP, GDP and Hadacidin | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1LOO
 
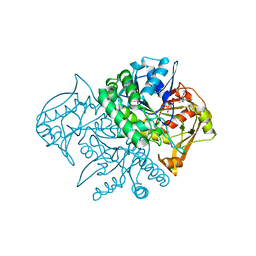 | | Crystal Structure of the Mouse-Muscle Adenylosuccinate Synthetase Ligated with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, adenylosuccinate synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1MEZ
 
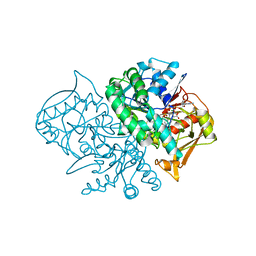 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with SAMP, GDP, SO4(2-), and Mg(2+) | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1MF0
 
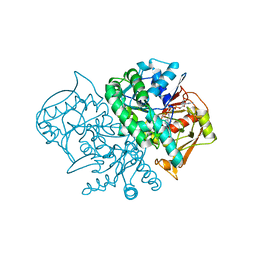 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP, GDP, HPO4(2-), and Mg(2+) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1MF1
 
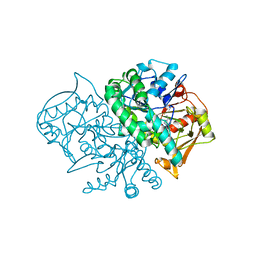 | | Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Adenylosuccinate Synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-08-09 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Feedback inhibition and product complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
4KXP
 
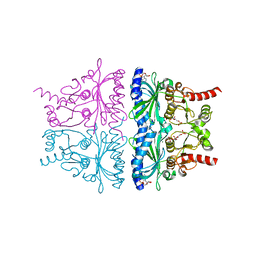 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant I10D in T-state | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2013-05-27 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Displacement of a Catalytically Essential Loop from the Active Site of Mammalian Fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
4LDS
 
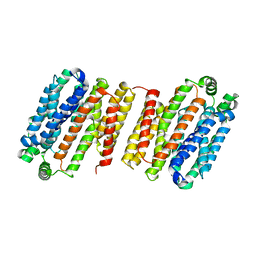 | |
1FJ9
 
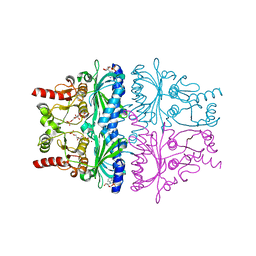 | | FRUCTOSE-1,6-BISPHOSPHATASE (MUTANT Y57W) PRODUCTS/ZN/AMP COMPLEX (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Iancu, C.V, Choe, J.Y, Honzatko, R.B. | | Deposit date: | 2000-08-07 | | Release date: | 2000-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tryptophan fluorescence reveals the conformational state of a dynamic loop in recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry, 39, 2000
|
|
1FJ6
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE (MUTANT Y57W) PRODUCT/ZN COMPLEX (R-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE-1,6-BISPHOSPHATASE, PHOSPHATE ION, ... | | Authors: | Iancu, C.V, Choe, J.Y, Honzatko, R.B. | | Deposit date: | 2000-08-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tryptophan fluorescence reveals the conformational state of a dynamic loop in recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry, 39, 2000
|
|
2F3B
 
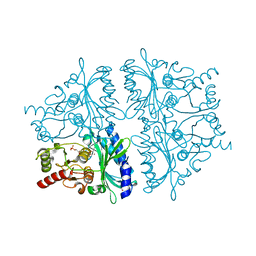 | | Mechanism of displacement of a catalytically essential loop from the active site of fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, PHOSPHATE ION, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-11-20 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
2F3D
 
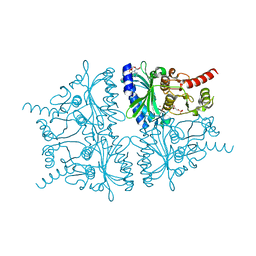 | | Mechanism of displacement of a catalytically essential loop from the active site of fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanism of displacement of a catalytically essential loop from the active site of mammalian fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
7JMR
 
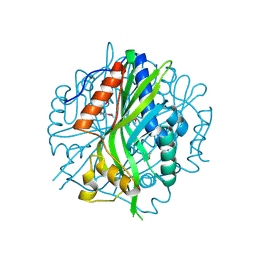 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pea pathogenicity protein 2 | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7JMV
 
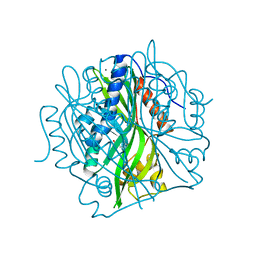 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis complexed with 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7KD9
 
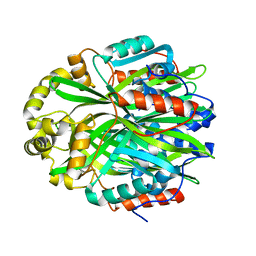 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | Gallate decarboxylase, POTASSIUM ION | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
6W54
 
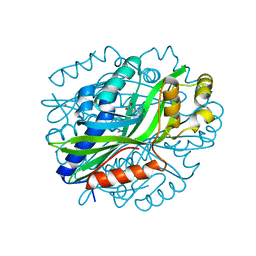 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | 4-NITROCATECHOL, COBALT (II) ION, Gallate decarboxylase, ... | | Authors: | Zeug, M, Marckovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
5U6M
 
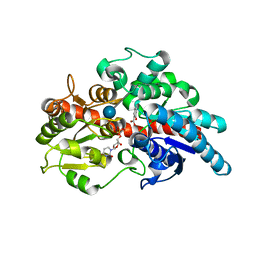 | | Crystal structure of UDP-glucosyltransferase, UGT74F2, with UDP and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, UDP-glycosyltransferase 74F2, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
5U6S
 
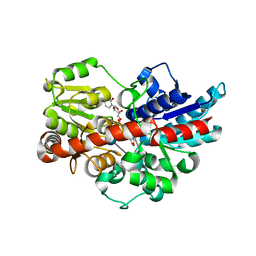 | | Crystal structure of UDP-glucosyltransferase, UGT74F2, with UDP and 2-bromobenzoic acid | | Descriptor: | 2-bromobenzoic acid, UDP-glycosyltransferase 74F2, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
5U6N
 
 | | Crystal structure of UDP-glucosyltransferase, UGT74F2 (T15S), with UDP and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, GLUTAMINE, UDP-glycosyltransferase 74F2, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
