6N60
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Microcin J25 (MccJ25) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4G0X
 
 | | Crystal Structure of Arabidopsis thaliana AGO1 MID domain | | Descriptor: | Protein argonaute 1, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2012-09-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
6N62
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N61
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Capistruin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capistruin, ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4G0P
 
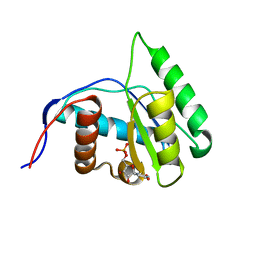 | | Crystal Structure of Arabidopsis thaliana AGO1 MID domain in complex with UMP | | Descriptor: | Protein argonaute 1, URIDINE-5'-MONOPHOSPHATE | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Y
 
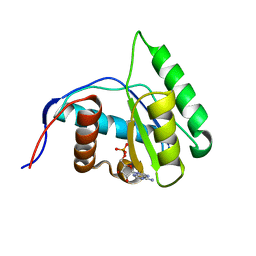 | | Crystal structure of Arabidopsis AGO1 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0O
 
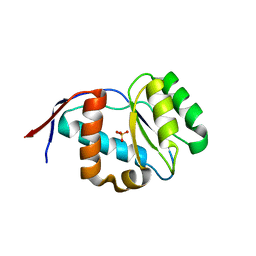 | | Crystal structure of Arabidopsis thaliana AGO5 MID domain | | Descriptor: | Protein argonaute 5, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Z
 
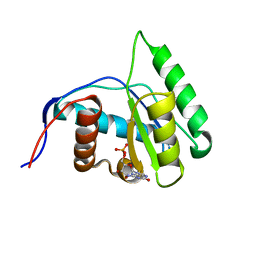 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0M
 
 | | Crystal structure of Arabidopsis thaliana AGO2 MID domain | | Descriptor: | Protein argonaute 2, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Q
 
 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
5TJG
 
 | |
