6CNC
 
 | | Yeast RNA polymerase III open complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNF
 
 | | Yeast RNA polymerase III elongation complex | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
5W5Y
 
 | | RNA polymerase I Initial Transcribing Complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W66
 
 | | RNA polymerase I Initial Transcribing Complex State 3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
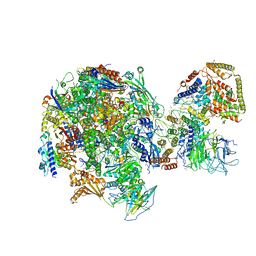 | | RNA Polymerase I Initial Transcribing Complex State 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W65
 
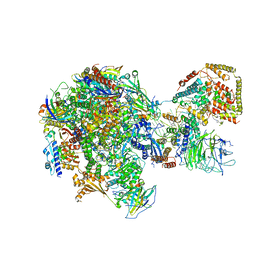 | | RNA polymerase I Initial Transcribing Complex State 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
4G8K
 
 | |
4G8L
 
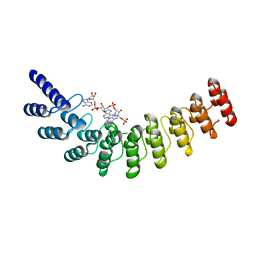 | | Active state of intact sensor domain of human RNase L with 2-5A bound | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Han, Y, Whitney, G, Donovan, J, Korennykh, A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Innate Immune Messenger 2-5A Tethers Human RNase L into Active High-Order Complexes.
Cell Rep, 2, 2012
|
|
4OAV
 
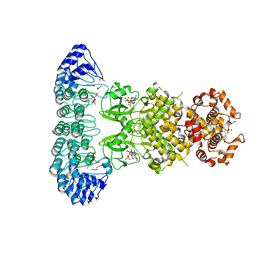 | | Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. | | Descriptor: | (2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxy-2-({[(S)-hydroxy{[(2R,3S,4S)-4-hydroxy-2-(hydroxymethyl)tetrahydrofuran-3-yl]oxy}phosphoryl]oxy}methyl)tetrahydrofuran-3-yl dihydrogen phosphate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
4OAU
 
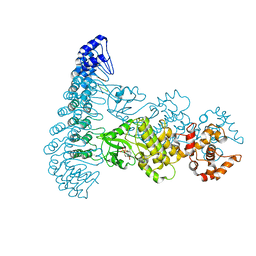 | | Complete human RNase L in complex with biological activators. | | Descriptor: | 2-5A-dependent ribonuclease, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
6CND
 
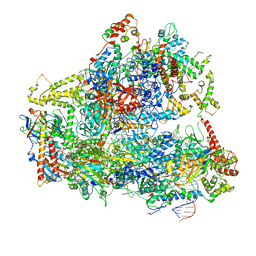 | | Yeast RNA polymerase III natural open complex (nOC) | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNB
 
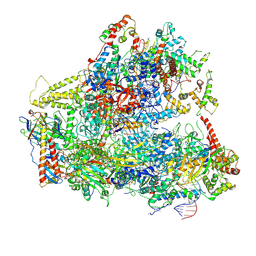 | | Yeast RNA polymerase III initial transcribing complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
8XRY
 
 | |
8XS5
 
 | |
8XVX
 
 | |
8XW2
 
 | |
8XVY
 
 | |
8XW3
 
 | |
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XS4
 
 | |
8XNG
 
 | |
8XVZ
 
 | |
8XS0
 
 | |
8XW0
 
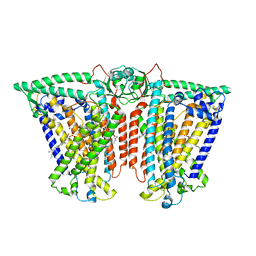 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
8XW1
 
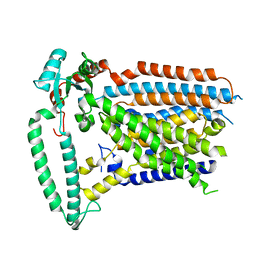 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 2024
|
|
