2FB3
 
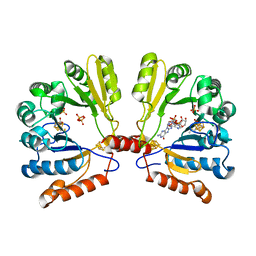 | | Structure of MoaA in complex with 5'-GTP | | Descriptor: | 5'-DEOXYADENOSINE, GUANOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FB2
 
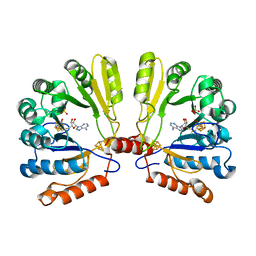 | | Structure of the MoaA Arg17/266/268/Ala triple mutant | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, S-ADENOSYLMETHIONINE, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3PVJ
 
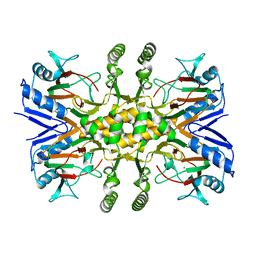 | | Crystal structure of the Fe(II)/alpha-ketoglutarate dependent taurine dioxygenase from Pseudomonas putida KT2440 | | Descriptor: | Alpha-ketoglutarate-dependent taurine dioxygenase, CHLORIDE ION | | Authors: | Knauer, S.H, Spiegelhauer, O, Schwarzinger, S, Haenzelmann, P, Dobbek, H. | | Deposit date: | 2010-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of the Fe(II)/alpha-ketoglutarate dependent taurine dioxygenase from Pseudomonas putida KT2440
To be Published
|
|
3TIW
 
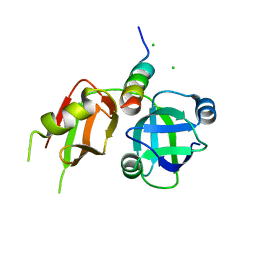 | | Crystal structure of p97N in complex with the C-terminus of gp78 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase AMFR, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Structural and Functional Basis of the p97/Valosin-containing Protein (VCP)-interacting Motif (VIM): MUTUALLY EXCLUSIVE BINDING OF COFACTORS TO THE N-TERMINAL DOMAIN OF p97.
J.Biol.Chem., 286, 2011
|
|
1TV7
 
 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TV8
 
 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1FFU
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
1FFV
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
3IPP
 
 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3M63
 
 | |
3M62
 
 | |
3QQ7
 
 | |
3QQ8
 
 | |
5C18
 
 | | p97-delta709-728 in complex with ATP-gamma-S | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
5C19
 
 | | p97 variant 2 in the apo state | | Descriptor: | SULFATE ION, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
5C1B
 
 | |
5C1A
 
 | | p97-N750D/R753D/M757D/Q760D in complex with ATP-gamma-S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97.
Structure, 24, 2016
|
|
3R3M
 
 | |
2VSF
 
 | | Structure of XPD from Thermoplasma acidophilum | | Descriptor: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
