4AJV
 
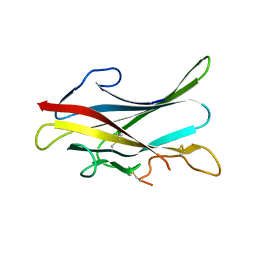 | | Structure of mouse ZP-C domain of TGF-Beta-Receptor-3 | | Descriptor: | TRANSFORMING GROWTH FACTOR BETA RECEPTOR TYPE 3 | | Authors: | Diestel, U, Resch, M, Meinhardt, K, Weiler, S, Hellmann, T.V, Nickel, J, Eichler, J, Muller, Y.A. | | Deposit date: | 2012-02-20 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a Novel Tgf-Beta-Binding Site in the Zona Pellucida C-Terminal (Zp-C) Domain of Tgf-Beta-Receptor-3 (Tgfr-3).
Plos One, 8, 2013
|
|
6TVQ
 
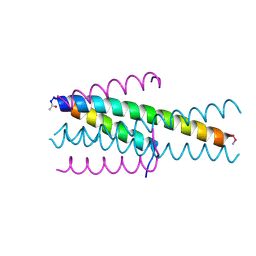 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVU
 
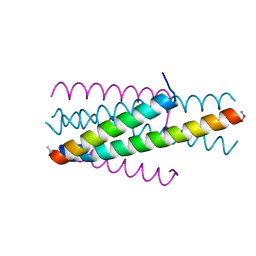 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Transmembrane protein gp41 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVW
 
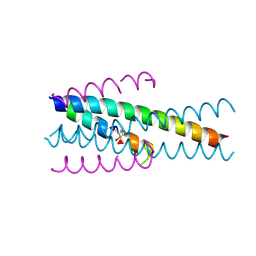 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | (2~{S},3~{S})-2-azanyl-5,5,5-tris(fluoranyl)-3-methyl-pentanal, ASN-ASN-TYR-THR-SER-LEU-ILE-HIS-SER-LEU-ILE-GLU-GLU, Envelope glycoprotein, ... | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | Descriptor: | pGolemi | | Authors: | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | Deposit date: | 2008-08-03 | | Release date: | 2009-06-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
4NCV
 
 | | Foldon domain wild type N-conjugate | | Descriptor: | Fibritin | | Authors: | Graewert, M.A, Berthelmann, A, Lach, J, Groll, M, Eichler, J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
4NCW
 
 | | foldon domain wild type C-conjugate | | Descriptor: | Fibritin, N,N',N''-triethylbenzene-1,3,5-tricarboxamide | | Authors: | Graewert, M.A, Berthelmann, A, Lach, J, Groll, M, Eichler, J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
7PAB
 
 | | Varicella zoster Orf24-Orf27 nuclear egress complex | | Descriptor: | Nuclear egress protein 2,Nuclear egress protein 1, SULFATE ION, ZINC ION | | Authors: | Schweininger, J, Muller, Y.A. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the varicella-zoster Orf24-Orf27 nuclear egress complex spotlights multiple determinants of herpesvirus subfamily specificity.
J.Biol.Chem., 298, 2022
|
|
6EZ1
 
 | |
6F5C
 
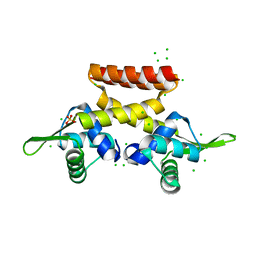 | |
6FDH
 
 | |
6FAQ
 
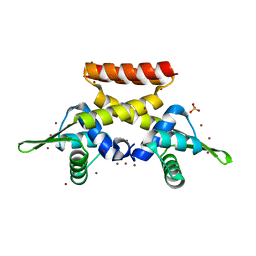 | |
4NCU
 
 | | Foldon domain wild type | | Descriptor: | Fibritin | | Authors: | Graewert, M.A, Reiner, A, Groll, M, Kiefhaber, T. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
6T3X
 
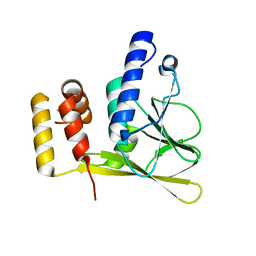 | | Crystal structure of the truncated human cytomegalovirus pUL50-pUL53 complex | | Descriptor: | Nuclear egress protein 2,Nuclear egress protein 1 | | Authors: | Muller, Y.A. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity.
J.Biol.Chem., 295, 2020
|
|
7NFB
 
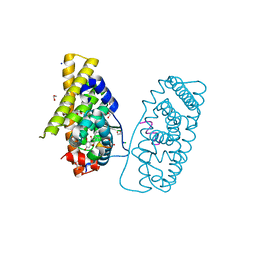 | |
7NEL
 
 | |
7NDO
 
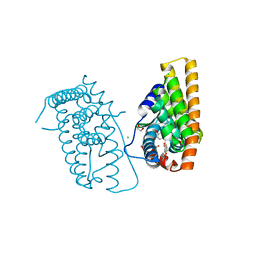 | | ER-PRS*(-) (L536S, L372R) in complex with raloxifene | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Estrogen receptor, ... | | Authors: | Kriegel, M, Muller, Y.A. | | Deposit date: | 2021-02-02 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A PROSS-designed extensively mutated estrogen receptor alpha variant displays enhanced thermal stability while retaining native allosteric regulation and structure.
Sci Rep, 11, 2021
|
|
6T3Z
 
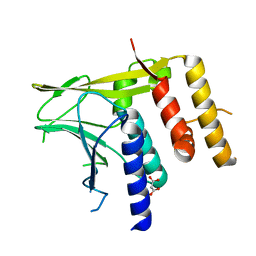 | | Crystal structure of the truncated EBV BFRF1-BFLF2 nuclear egress complex | | Descriptor: | MALONATE ION, Nuclear egress protein 2,Nuclear egress protein 1 | | Authors: | Muller, Y.A. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55862784 Å) | | Cite: | High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity.
J.Biol.Chem., 295, 2020
|
|
6QFD
 
 | | The complex structure of hsRosR-S4 (vng0258/RosR-S4) | | Descriptor: | DNA (28-MER), DNA-binding protein, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QH0
 
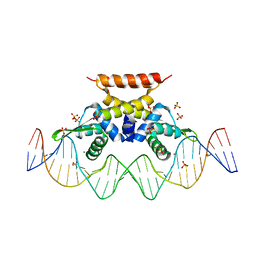 | | The complex structure of hsRosR-S5 (VNG0258H/RosR-S5) | | Descriptor: | DNA (28-MER), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QIL
 
 | | The complex structure of hsRosR-S1 (VNG0258H/RosR-S1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (28-MER), DNA binding protein, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QUA
 
 | | The complex structure of hsRosR-SG (vng0258/RosR-SG) | | Descriptor: | DNA (28-MER), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
