3CRD
 
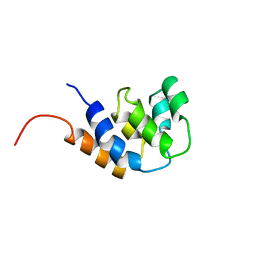 | | NMR STRUCTURE OF THE RAIDD CARD DOMAIN, 15 STRUCTURES | | Descriptor: | RAIDD | | Authors: | Chou, J.J, Matsuo, H, Duan, H, Wagner, G. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment.
Cell(Cambridge,Mass.), 94, 1998
|
|
8TE6
 
 | |
8TE5
 
 | |
4X0G
 
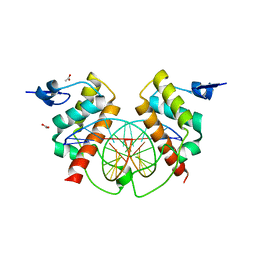 | | Structure of Bsg25A binding with DNA | | Descriptor: | ACETATE ION, Blastoderm-specific gene 25A, DNA (5'-D(*GP*TP*TP*CP*CP*AP*AP*TP*TP*GP*GP*AP*A)-3') | | Authors: | Ren, A. | | Deposit date: | 2014-11-21 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2065 Å) | | Cite: | Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family.
Genes Dev., 29, 2015
|
|
8TYP
 
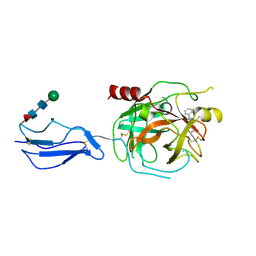 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
6MQE
 
 | |
6MQS
 
 | |
6NF2
 
 | |
6OT1
 
 | |
6OSY
 
 | |
4HZL
 
 | |
8F7T
 
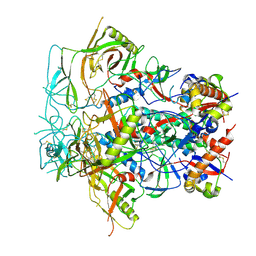 | | Glycan-Base ConC Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env gp120, ... | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-20 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases.
Iscience, 26, 2023
|
|
6MPH
 
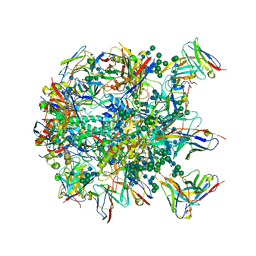 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MQC
 
 | |
6MQM
 
 | |
6MQR
 
 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6N16
 
 | |
6N1V
 
 | |
6N1W
 
 | |
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
7E26
 
 | | Structure of PfFNT in apo state | | Descriptor: | Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7E27
 
 | | Structure of PfFNT in complex with MMV007839 | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7JKS
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
6LYN
 
 | | CD146 D4-D5/AA98 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AA98 Fab heavy chain, ... | | Authors: | Chen, X, Yan, X. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structure basis for AA98 inhibition on the activation of endothelial cells mediated by CD146.
Iscience, 24, 2021
|
|
