5C2X
 
 | | Crystal structure of deoxyribose-phosphate aldolase from Colwellia psychrerythraea (tetragonal form) | | Descriptor: | CARBONATE ION, Deoxyribose-phosphate aldolase, SULFATE ION, ... | | Authors: | Dick, M, Weiergraeber, O.H, Pietruszka, J. | | Deposit date: | 2015-06-16 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Trading off stability against activity in extremophilic aldolases.
Sci Rep, 6, 2016
|
|
5EKY
 
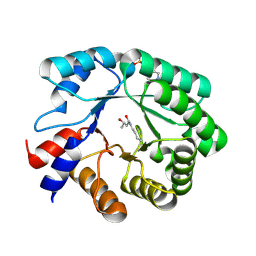 | | Crystal structure of deoxyribose-phosphate aldolase from Escherichia coli (K58E-Y96W mutant) | | Descriptor: | 1,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Deoxyribose-phosphate aldolase | | Authors: | Classen, T, Dick, M, Pietruszka, J, Weiergraeber, O.H. | | Deposit date: | 2015-11-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: structural insights and protective strategies.
Chem Sci, 7, 2016
|
|
5EMU
 
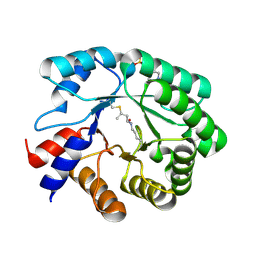 | |
5EL1
 
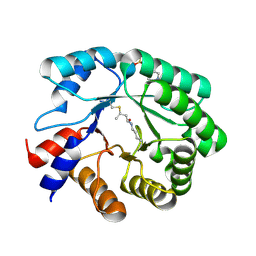 | |
5C6M
 
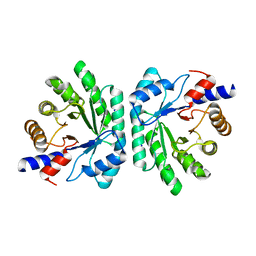 | | Crystal structure of deoxyribose-phosphate aldolase from Shewanella halifaxensis | | Descriptor: | CHLORIDE ION, Deoxyribose-phosphate aldolase, SODIUM ION | | Authors: | Weiergraeber, O.H, Dick, M, Bramski, J, Pietruszka, J. | | Deposit date: | 2015-06-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Trading off stability against activity in extremophilic aldolases.
Sci Rep, 6, 2016
|
|
5C5Y
 
 | |
2N1F
 
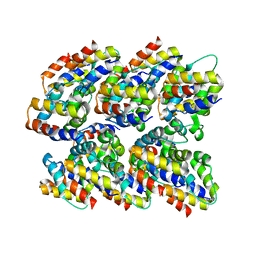 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6GBE
 
 | |
6GBD
 
 | |
