1MFN
 
 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 20 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
2MFN
 
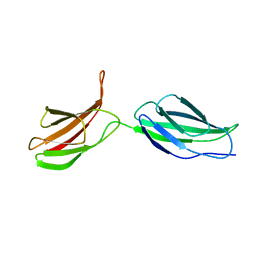 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
4AAI
 
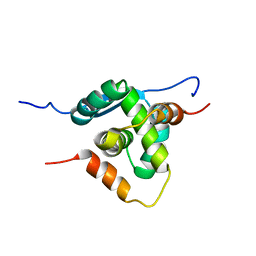 | | THERMOSTABLE PROTEIN FROM HYPERTHERMOPHILIC VIRUS SSV-RH | | Descriptor: | ORF E73 | | Authors: | Schlenker, C, Goel, A, Tripet, B.P, Menon, S, Lawrence, C.M, Copie, V. | | Deposit date: | 2011-12-02 | | Release date: | 2012-01-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of E73 from a Hyperthermophilic Archaeal Virus Identify the "Rh3" Domain, an Elaborated Ribbon-Helix- Helix Motif Involved in DNA Recognition.
Biochemistry, 51, 2012
|
|
2MPU
 
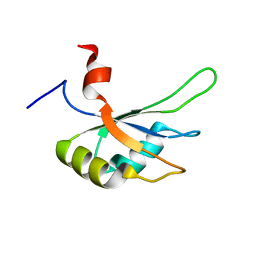 | | Structural and Functional analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich RNA binding protein implicated in the regulation of barley leaf senescence and environmental adaptation | | Descriptor: | RBP1 | | Authors: | Mason, K.E, Tripet, B.P, Eilers, B.J, Powell, P, Fischer, A.M, Copie, V. | | Deposit date: | 2014-06-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Analysis of the Hordeum vulgare L. HvGR-RBP1 Protein, a Glycine-Rich RNA-Binding Protein Involved in the Regulation of Barley Plant Development and Stress Response.
Biochemistry, 53, 2014
|
|
2MOQ
 
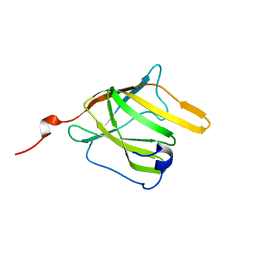 | | Solution Structure and Molecular determinants of Hemoglobin Binding of the first NEAT Domain of IsdB in Staphylococcus aureus | | Descriptor: | Iron-regulated surface determinant protein B | | Authors: | Fonner, B.A, Tripet, B.P, Eilers, B.J, Stanisich, J, Sullivan-Springhetti, R.K, Moore, R, Lui, M, Lei, B, Copie, V. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Molecular Determinants of Hemoglobin Binding of the First NEAT Domain of IsdB in Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
2WTE
 
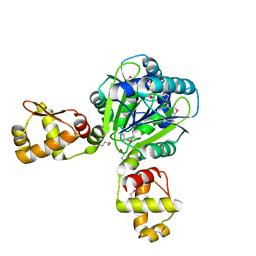 | | The structure of the CRISPR-associated protein, Csa3, from Sulfolobus solfataricus at 1.8 angstrom resolution. | | Descriptor: | CSA3, DI(HYDROXYETHYL)ETHER | | Authors: | Lintner, N.G, Alsbury, D.L, Copie, V, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Crispr-Associated Protein Csa3 Provides Insight Into the Regulation of the Crispr/Cas System.
J.Mol.Biol., 405, 2011
|
|
2XDI
 
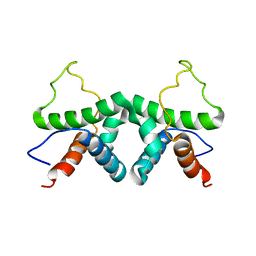 | |
1XI7
 
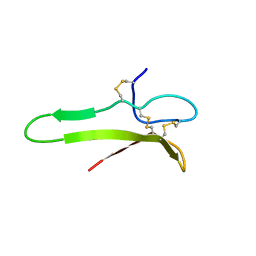 | | NMR structure of the carboxyl-terminal cysteine domain of the VHv1.1 polydnaviral gene product | | Descriptor: | cysteine-rich omega-conotoxin homolog VHv1.1 | | Authors: | Einerwold, J, Jaseja, M, Hapner, K, Webb, B, Copie, V. | | Deposit date: | 2004-09-21 | | Release date: | 2004-10-05 | | Last modified: | 2011-08-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carboxyl-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product: comparison with other cystine knot structural folds
Biochemistry, 40, 2001
|
|
1XJ1
 
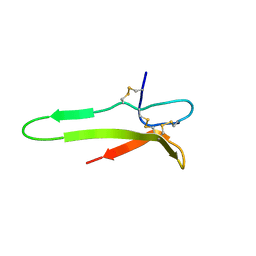 | | 3D solution structure of the C-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product | | Descriptor: | cysteine-rich omega-conotoxin homolog VHv1.1 | | Authors: | Einerwold, J, Jaseja, J, Hapner, K, Webb, B, Copie, V. | | Deposit date: | 2004-09-22 | | Release date: | 2004-10-05 | | Last modified: | 2011-08-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carboxyl-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product: comparison with other cystine knot structural folds
Biochemistry, 40, 2001
|
|
2KKG
 
 | |
2HPU
 
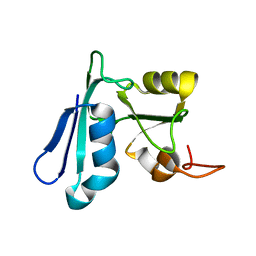 | | Solution NMR structure of the apo-NosL protein from Achromobacter cycloclastes | | Descriptor: | NosL protein | | Authors: | Taubner, L.M, McGuirl, M.A, Dooley, D.M, Copie, V. | | Deposit date: | 2006-07-17 | | Release date: | 2006-10-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Apo Nosl, an Accessory Protein of the Nitrous Oxide Reductase System: Insights from Structural Homology with MerB, a Mercury Resistance Protein.
Biochemistry, 45, 2006
|
|
2HQ3
 
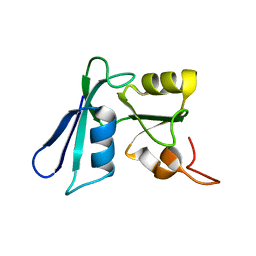 | | Solution NMR structure of the apo-NosL protein from Achromobacter cycloclastes | | Descriptor: | NosL protein | | Authors: | Taubner, L.M, McGuirl, M.A, Dooley, D.M, Copie, V. | | Deposit date: | 2006-07-18 | | Release date: | 2006-10-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Apo Nosl, an Accessory Protein of the Nitrous Oxide Reductase System: Insights from Structural Homology with MerB, a Mercury Resistance Protein.
Biochemistry, 45, 2006
|
|
3PS0
 
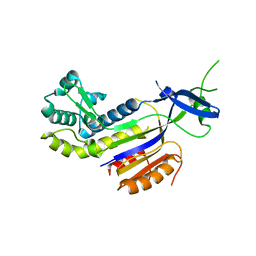 | | The structure of the CRISPR-associated protein, csa2, from Sulfolobus solfataricus | | Descriptor: | CRISPR-Associated protein, CSA2 | | Authors: | Lintner, N.G, Sdano, M, Young, M.J, Lawrence, C.M. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE).
J.Biol.Chem., 286, 2011
|
|
6QZ2
 
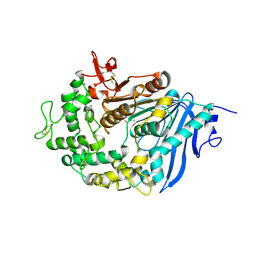 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QZ3
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QZ4
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, SULFATE ION | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QZ1
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
