1M9B
 
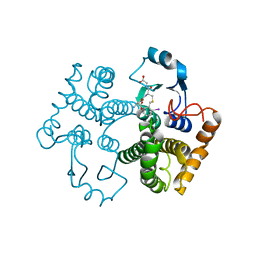 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with gamma-glutamyl[S-(2-iodobenzyl)cysteinyl]glycine | | Descriptor: | GAMMA-GLUTAMYL[S-(2-IODOBENZYL)CYSTEINYL]GLYCINE, Glutathione S-Transferase 26 kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M99
 
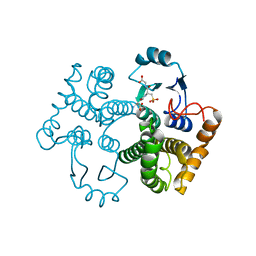 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with glutathione sulfonic acid | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-Transferase 26kDa | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M9A
 
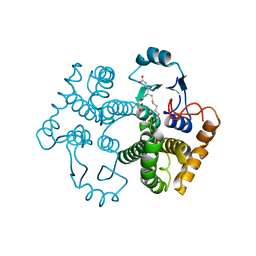 | | Crystal structure of the 26 kDa glutathione S-transferase from Schistosoma japonicum complexed with S-hexylglutathione | | Descriptor: | Glutathione S-Transferase 26 kDa, S-HEXYLGLUTATHIONE | | Authors: | Cardoso, R.M.F, Daniels, D.S, Bruns, C.M, Tainer, J.A. | | Deposit date: | 2002-07-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the electrophile
binding site and substrate binding
mode of the 26-kDa glutathione
S-transferase from Schistosoma
japonicum
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1TZG
 
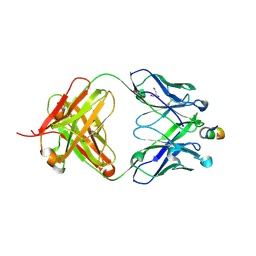 | | Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 | | Descriptor: | Envelope polyprotein GP160, Fab 4E10, GLYCEROL | | Authors: | Cardoso, R.M.F, Zwick, M.B, Stanfield, R.L, Kunert, R, Binley, J.M, Katinger, H, Burton, D.R, Wilson, I.A. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41
Immunity, 22, 2005
|
|
1N19
 
 | | Structure of the HSOD A4V mutant | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide Dismutase [Cu-Zn], ... | | Authors: | Cardoso, R.M.F, Thayer, M.M, DiDonato, M, Lo, T.P, Bruns, C.K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2002-10-16 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase.
J.Mol.Biol., 324, 2002
|
|
1N18
 
 | | Thermostable mutant of Human Superoxide Dismutase, C6A, C111S | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cardoso, R.M.F, Thayer, M.M, DiDonato, M, Lo, T.P, Bruns, C.K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2002-10-16 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase.
J.Mol.Biol., 324, 2002
|
|
6WVZ
 
 | | Crystal structure of anti-MET Fab arm of amivantamab in complex with human MET | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2020-05-07 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of amivantamab (JNJ-61186372), a bispecific antibody targeting EGFR and MET.
J.Biol.Chem., 296, 2021
|
|
2FX9
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a thioether-linked peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein gp41 | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
2FX8
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with an aib-induced peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41) | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
2FX7
 
 | | Crystal structure of hiv-1 neutralizing human fab 4e10 in complex with a 16-residue peptide encompassing the 4e10 epitope on gp41 | | Descriptor: | Fab 4E10, Fragment of HIV glycoprotein (GP41), GLYCEROL | | Authors: | Cardoso, R.M.F, Brunel, F.M, Ferguson, S, Burton, D.R, Dawson, P.E, Wilson, I.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10.
J.Mol.Biol., 365, 2007
|
|
1TO5
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase, ... | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TO4
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | COPPER (II) ION, Superoxide dismutase, ZINC ION | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PU0
 
 | | Structure of Human Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Craig, L, Huff, M.E, Thayer, M.M, Cardoso, R.M.F, Kassmann, C.J, Lo, T.P, Bruns, C.K, Powers, E.T, Kelly, J.W, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates Via Framework Destabilization
J.Mol.Biol., 332, 2003
|
|
1PTZ
 
 | | Crystal structure of the human CU, Zn Superoxide Dismutase, Familial Amyotrophic Lateral Sclerosis (FALS) Mutant H43R | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Craig, L, Huff, M.E, Thayer, M.M, Cardoso, R.M.F, Kassmann, C.J, Lo, T.P, Bruns, C.K, Powers, E.T, Kelly, J.W, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates Via Framework Destabilization
J.Mol.Biol., 332, 2003
|
|
5L2H
 
 | | Crystal Structure of W26A mutant of anti-EGFR Centyrin P54AR4-83v2 | | Descriptor: | Centyrin, GLYCEROL | | Authors: | Cardoso, R.M.F, Goldberg, S.D, O Neil, K.T, Gilliland, G.L. | | Deposit date: | 2016-08-01 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8013 Å) | | Cite: | Engineering a targeted delivery platform using Centyrins.
Protein Eng. Des. Sel., 29, 2016
|
|
5TZU
 
 | | Crystal structure of human CD47 ECD bound to Fab of B6H12.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
5TZ2
 
 | | Crystal structure of human CD47 ECD bound to Fab of C47B222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C47B222 Fab Heavy Chain, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
5TZT
 
 | | Crystal structure of human CD47 ECD bound to Fab of C47B161 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy Chain of Fab C47B161, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
