1O9W
 
 | | F17-aG lectin domain from Escherichia coli in complex with N-acetyl-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17A-G FIMBRIAL ADHESIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1O9V
 
 | | F17-aG lectin domain from Escherichia coli in complex with a selenium carbohydrate derivative | | Descriptor: | F17-AG LECTIN DOMAIN, methyl 2-acetamido-2-deoxy-1-seleno-beta-D-glucopyranoside | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1O9Z
 
 | | F17-aG lectin domain from Escherichia coli (ligand free) | | Descriptor: | F17-AG LECTIN DOMAIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-23 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
2CD7
 
 | | Staphylococcus aureus pI258 arsenate reductase (ArsC) H62Q mutant | | Descriptor: | PROTEIN ARSC, SODIUM ION | | Authors: | Buts, L, Roos, G, Van Belle, K, Brosens, E, Loris, R, Wyns, L, Messens, J. | | Deposit date: | 2006-01-23 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interplay between Ion Binding and Catalysis in the Thioredoxin-Coupled Arsenate Reductase Family.
J.Mol.Biol., 360, 2006
|
|
1LK0
 
 | | Disulfide intermediate of C89L Arsenate reductase from pI258 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, arsenate reductase | | Authors: | Messens, J, Martins, J.C, Van Belle, K, Brosens, E, Desmyter, A, De Gieter, M, Wieruszeski, J.M, Willem, R, Wyns, L, Zegers, I. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LJL
 
 | | Wild Type pI258 S. aureus arsenate reductase | | Descriptor: | POTASSIUM ION, arsenate reductase | | Authors: | Messens, J, Martins, J.C, Van Belle, K, Brosens, E, Desmyter, A, De Gieter, M, Wieruszeski, J.M, Willem, R, Wyns, L, Zegers, I. | | Deposit date: | 2002-04-21 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2O7K
 
 | | S. aureus thioredoxin | | Descriptor: | Thioredoxin | | Authors: | Roos, G, Garcia-Pino, A, Van Belle, K, Brosens, E, Wahni, K, Vandenbussche, G, Wyns, L, Loris, R, Messens, J. | | Deposit date: | 2006-12-11 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin
J.Mol.Biol., 368, 2007
|
|
2FXI
 
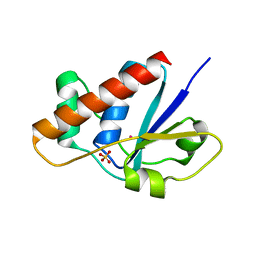 | |
2PQY
 
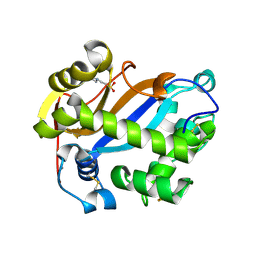 | | E. coli RNase 1 (in vitro refolded with DsbA only) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Ribonuclease I | | Authors: | Messens, J, Loris, R. | | Deposit date: | 2007-05-03 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The oxidase DsbA folds a protein with a nonconsecutive disulfide.
J.Biol.Chem., 282, 2007
|
|
1YOE
 
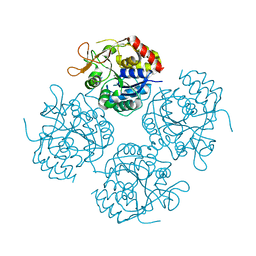 | |
1HOZ
 
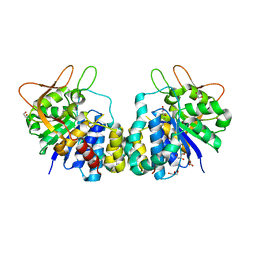 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX | | Descriptor: | CALCIUM ION, GLYCEROL, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1LJU
 
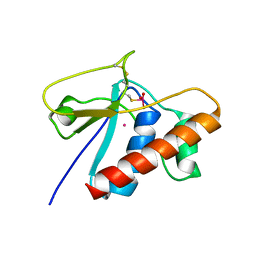 | | X-RAY STRUCTURE OF C15A ARSENATE REDUCTASE FROM PI258 COMPLEXED WITH ARSENITE | | Descriptor: | POTASSIUM ION, arsenate reductase | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2002-04-22 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2O89
 
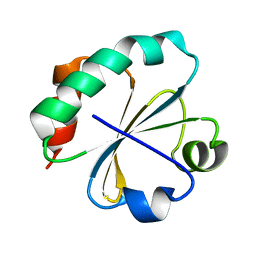 | | S. aureus thioredoxin P31T/C32S mutant | | Descriptor: | Thioredoxin | | Authors: | Roos, G, Loris, R, Messens, J. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin
J.Mol.Biol., 368, 2007
|
|
2O85
 
 | | S. Aureus thioredoxin P31T mutant | | Descriptor: | Thioredoxin | | Authors: | Roos, G, Loris, R, Messens, J. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin
J.Mol.Biol., 368, 2007
|
|
2O87
 
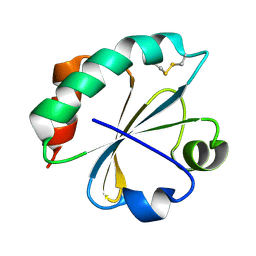 | | S. aureus thioredoxin P31S mutant | | Descriptor: | Thioredoxin | | Authors: | Roos, G, Loris, R, Messens, J. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The conserved active site proline determines the reducing power of Staphylococcus aureus thioredoxin
J.Mol.Biol., 368, 2007
|
|
1Q8F
 
 | |
