4U87
 
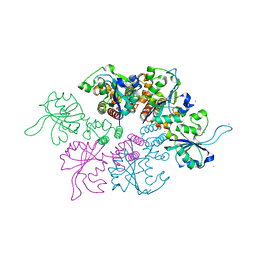 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | Descriptor: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
3I1D
 
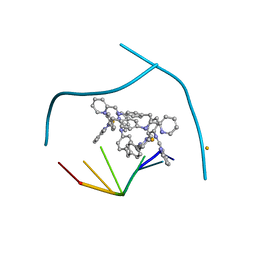 | | Distinct recognition of three-way DNA junctions by the two enantiomers of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Boer, D.R, Uson, I, Hannon, M.J, Coll, M. | | Deposit date: | 2009-06-26 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3FX8
 
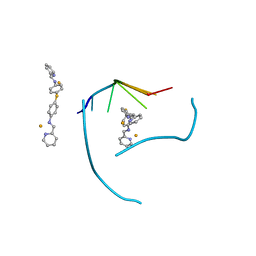 | | Distinct recognition of three-way DNA junctions by a thioester variant of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | (5'-D(*CP*GP*TP*AP*CP*G)-3', 4,4'-sulfanediylbis{N-[(1E)-pyridin-2-ylmethylidene]aniline}, FE (II) ION | | Authors: | Boer, D.R, Uson, I, Coll, M. | | Deposit date: | 2009-01-20 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
4LDX
 
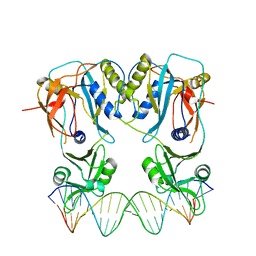 | | Crystal structure of the DNA binding domain of arabidopsis thaliana auxin response factor 1 (ARF1) in complex with protomor-like sequence ER7 | | Descriptor: | Auxin response factor 1, ER7, forward sequence, ... | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
1SIJ
 
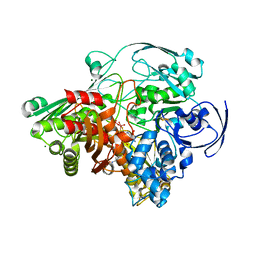 | | Crystal structure of the Aldehyde Dehydrogenase (a.k.a. AOR or MOP) of Desulfovibrio gigas covalently bound to [AsO3]- | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ARSENITE, Aldehyde oxidoreductase, ... | | Authors: | Boer, D.R, Thapper, A, Brondino, C.D, Romao, M.J, Moura, J.J.G. | | Deposit date: | 2004-03-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure and EPR Spectra of "Arsenite-Inhibited" Desulfovibriogigas Aldehyde Dehydrogenase: A Member of the Xanthine Oxidase Family
J.Am.Chem.Soc., 126, 2004
|
|
4GQJ
 
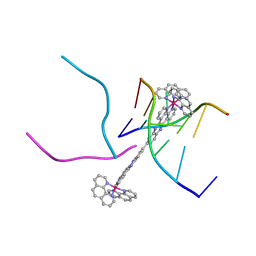 | | Complex of a binuclear Ruthenium compound D,D-([mu-(11,11')-bi(dppz)-(1,10-phenanthroline)4-Ru2]4+) bound to d(CGTACG) | | Descriptor: | (mu-11,11'-bidipyrido[3,2-a:2',3'-c]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~4'~,N~5'~)[tetrakis(1,10-phenanthroline-kappa~2~N~1~,N~10~)]diruthenium, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Boer, D.R, Coll, M. | | Deposit date: | 2012-08-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Thread Insertion of a Bis(dipyridophenazine) Diruthenium Complex into the DNA Double Helix by the Extrusion of AT Base Pairs and Cross-Linking of DNA Duplexes.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4LDU
 
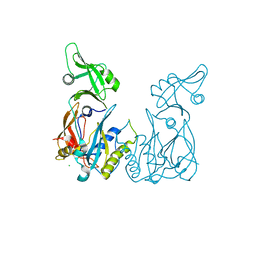 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana auxin response factor 5 | | Descriptor: | Auxin response factor 5, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDW
 
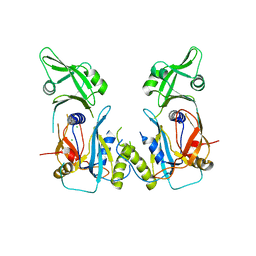 | |
4LDY
 
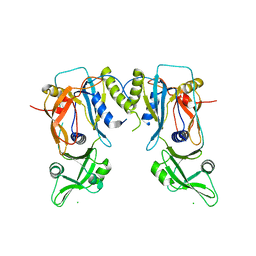 | | Crystal structure of the DNA binding domain of the G245A mutant of arabidopsis thaliana auxin reponse factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDV
 
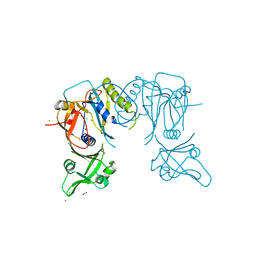 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
3DKX
 
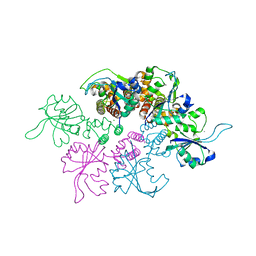 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), trigonal form, to 2.7 Ang resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
3DKY
 
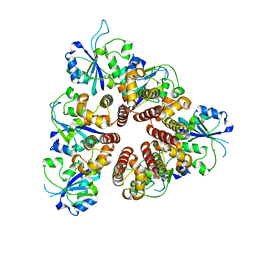 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
3L4P
 
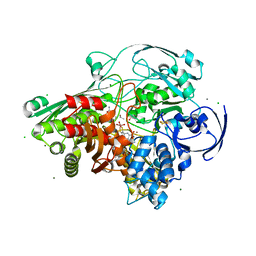 | |
7QNQ
 
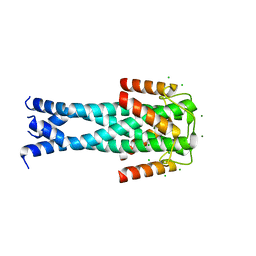 | | Structure of the Aux2 relaxosome protein of plasmid pLS20 | | Descriptor: | Auxiliary relaxosome protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Boer, D.R, Crespo, I. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical characterization of the relaxosome auxiliary proteins encoded on the Bacillus subtilis plasmid pLS20.
Comput Struct Biotechnol J, 20, 2022
|
|
8OJ1
 
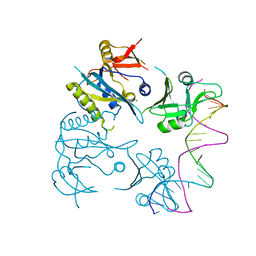 | |
8OJ2
 
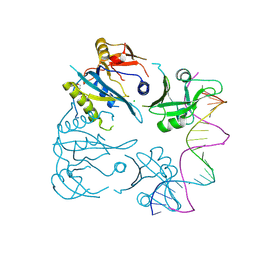 | |
5N2Q
 
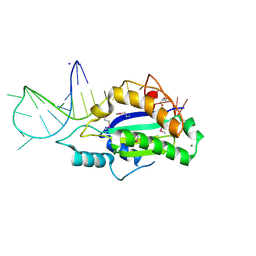 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to 26nt pMV158 oriT DNA | | Descriptor: | CHLORIDE ION, DNA (26-MER), GLYCEROL, ... | | Authors: | Russi, S, Boer, D.R, Coll, M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
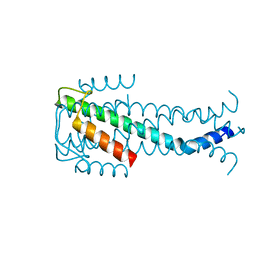
7NUV
 
 | |
8BNY
 
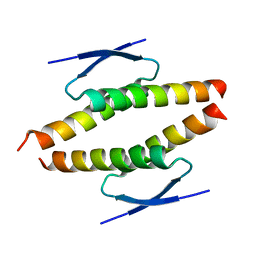 | | Structure of the tetramerization domain of pLS20 conjugation repressor Rco | | Descriptor: | CHLORIDE ION, Immunity repressor protein | | Authors: | Bernardo, N, Crespo, I, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | A tetramerization domain in prokaryotic and eukaryotic transcription regulators homologous to p53.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6SDG
 
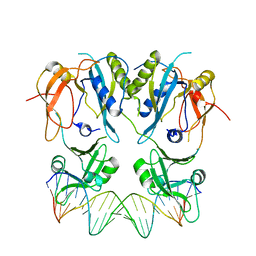 | |
6GYG
 
 | |
7BCB
 
 | |
7BCA
 
 | |
7BBQ
 
 | |
6YCQ
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana Auxin Response Factor 1 (AtARF1) in complex with High Affinity DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 21-7A, 21-7B, ... | | Authors: | Crespo, I, Weijers, D, Boer, D.R. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
