1B3Q
 
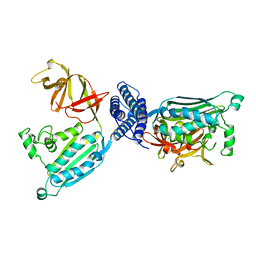 | | CRYSTAL STRUCTURE OF CHEA-289, A SIGNAL TRANSDUCING HISTIDINE KINASE | | Descriptor: | MERCURY (II) ION, PROTEIN (CHEMOTAXIS PROTEIN CHEA) | | Authors: | Bilwes, A.M, Alex, L.A, Crane, B.R, Simon, M.I. | | Deposit date: | 1998-12-14 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of CheA, a signal-transducing histidine kinase.
Cell(Cambridge,Mass.), 96, 1999
|
|
1I5C
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5B
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPNP AND MANGANESE | | Descriptor: | ACETATE ION, CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I59
 
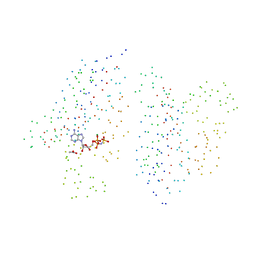 | | STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ADPNP AND MAGENSIUM | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1YFO
 
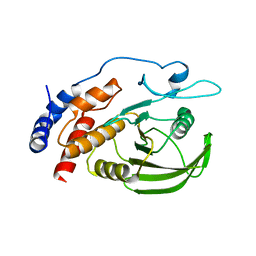 | |
1I58
 
 | | STRUCTURE OF THE HISTIDINE KINASE CHEA ATP-BINDING DOMAIN IN COMPLEX WITH ATP ANALOG ADPCP AND MAGNESIUM | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHEMOTAXIS PROTEIN CHEA, ... | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5A
 
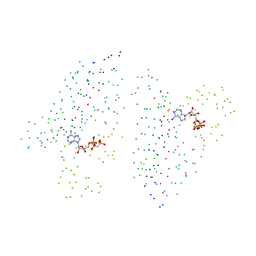 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH ADPCP AND MANGANESE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
1I5D
 
 | | STRUCTURE OF CHEA DOMAIN P4 IN COMPLEX WITH TNP-ATP | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, SULFATE ION | | Authors: | Bilwes, A.M, Quezada, C.M, Croal, L.R, Crane, B.R, Simon, M.I. | | Deposit date: | 2001-02-26 | | Release date: | 2001-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nucleotide binding by the histidine kinase CheA.
Nat.Struct.Biol., 8, 2001
|
|
6CNY
 
 | | 2.3 Angstrom Structure of Phosphodiesterase treated Vivid (complex with FMN) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Vivid PAS protein VVD | | Authors: | Zoltowski, B.D, Shabalin, I.G, Kowiel, M, Porebski, P.J, Crane, B.R, Bilwes, A.M. | | Deposit date: | 2018-03-09 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational switching in the fungal light sensor Vivid.
Science, 316, 2007
|
|
2HP7
 
 | | Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor | | Descriptor: | Flagellar motor switch protein FliM | | Authors: | Crane, B.R, Park, S, Lowder, B, Bilwes, A.M, Blair, D.F. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CH7
 
 | |
2CH4
 
 | |
2F9Z
 
 | | Complex between the chemotaxis deamidase CheD and the chemotaxis phosphatase CheC from Thermotoga maritima | | Descriptor: | PROTEIN (chemotaxis methylation protein), chemotaxis protein CheC | | Authors: | Chao, X, Park, S.Y, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation.
Cell(Cambridge,Mass.), 124, 2006
|
|
3LNR
 
 | |
3G67
 
 | |
3G6B
 
 | |
3SOH
 
 | | Architecture of the Flagellar Rotor | | Descriptor: | Flagellar motor switch protein FliG, Flagellar motor switch protein FliM | | Authors: | Koushik, P, Gonzalez-Bonet, G, Bilwes, A.M, Crane, B.R, Blair, D. | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the flagellar rotor.
Embo J., 30, 2011
|
|
3UR1
 
 | |
1U0S
 
 | | Chemotaxis kinase CheA P2 domain in complex with response regulator CheY from the thermophile thermotoga maritima | | Descriptor: | Chemotaxis protein cheA, Chemotaxis protein cheY | | Authors: | Park, S.Y, Beel, B.D, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XKR
 
 | | X-ray Structure of Thermotoga maritima CheC | | Descriptor: | chemotaxis protein CheC | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX
Mol.Cell, 16, 2004
|
|
1XKO
 
 | | Structure of Thermotoga maritima CheX | | Descriptor: | CHEMOTAXIS protein cheX | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX.
Mol.Cell, 16, 2004
|
|
2PD8
 
 | |
2PD7
 
 | |
2PDR
 
 | |
1TQG
 
 | | CheA phosphotransferase domain from Thermotoga maritima | | Descriptor: | Chemotaxis protein cheA | | Authors: | Quezada, C.M, Gradinaru, C, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima.
J.Mol.Biol., 341, 2004
|
|
