3RWN
 
 | |
4LIN
 
 | |
3LWW
 
 | |
5BU5
 
 | |
5BVZ
 
 | |
6PJX
 
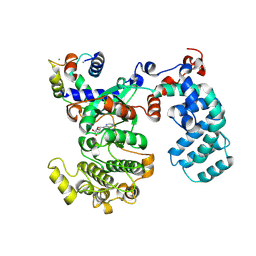 | | Crystal Structure of G Protein-Coupled Receptor Kinase 5 (GRK5) in Complex with Calmodulin (CaM) | | Descriptor: | CALCIUM ION, Calmodulin, G protein-coupled receptor kinase 5, ... | | Authors: | Bhardwaj, A, Komolov, K.E, Sulon, S, Benovic, J.L. | | Deposit date: | 2019-06-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a GRK5-Calmodulin Complex Reveals Molecular Mechanism of GRK Activation and Substrate Targeting.
Mol.Cell, 81, 2021
|
|
3P9A
 
 | |
4K6B
 
 | |
5KA5
 
 | |
4TND
 
 | |
4TNB
 
 | |
3Q5U
 
 | |
5KA6
 
 | |
2FGL
 
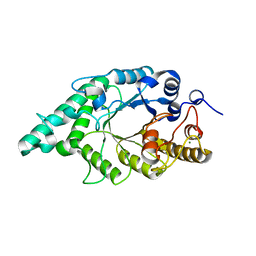 | | An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 | | Descriptor: | MAGNESIUM ION, alkaline thermostable endoxylanase, alpha-D-xylopyranose, ... | | Authors: | Ramakumar, S, Manikandan, K, Bhardwaj, A, Reddy, V.S, Lokanath, N.K, Ghosh, A. | | Deposit date: | 2005-12-22 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions.
Protein Sci., 15, 2006
|
|
2F8Q
 
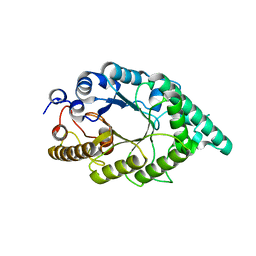 | | An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 | | Descriptor: | MAGNESIUM ION, alkaline thermostable endoxylanase | | Authors: | Ramakumar, S, Manikandan, K, Bhardwaj, A, Ghosh, A, Reddy, V.S. | | Deposit date: | 2005-12-03 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions.
Protein Sci., 15, 2006
|
|
4PVZ
 
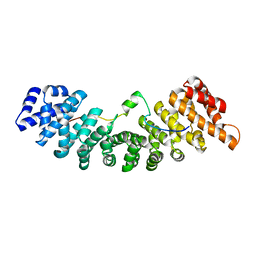 | | Structure of yeast importin a bound to the membrane protein Nuclear Localization Signal sequence of INM protein Heh2 | | Descriptor: | Importin subunit alpha, Inner nuclear membrane protein HEH2 | | Authors: | Lokareddy, R.K, Hapsari, A.R, van Rheenen, M, Bhardwaj, A, Veenhoff, L.M, Cingolani, C. | | Deposit date: | 2014-03-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive Properties of the Nuclear Localization Signals of Inner Nuclear Membrane Proteins Heh1 and Heh2.
Structure, 23, 2015
|
|
4HRF
 
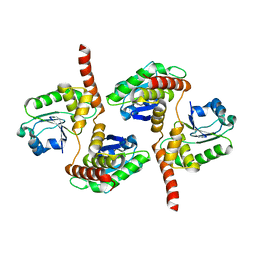 | | Atomic structure of DUSP26 | | Descriptor: | Dual specificity protein phosphatase 26 | | Authors: | Lokareddy, R.K, Bhardwaj, A, Cingolani, G. | | Deposit date: | 2012-10-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Atomic structure of dual-specificity phosphatase 26, a novel p53 phosphatase.
Biochemistry, 52, 2013
|
|
4QCE
 
 | | Crystal structure of recombinant alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, MAGNESIUM ION, SODIUM ION | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
4QCF
 
 | | Crystal structure of N-terminal mutant (V1A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
4QDM
 
 | | Crystal structure of N-terminal mutant (V1L) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-14 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
5EB8
 
 | |
5EBA
 
 | |
5EFF
 
 | | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Beta-xylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2015-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of an aromatic mutant (F4A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27
To Be Published
|
|
4ZKU
 
 | | P22 Tail Needle Gp26 crystallized at pH 10.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tail needle protein gp26 | | Authors: | Sankhala, R.S, Cingolani, G. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Plasticity of the Protein Plug That Traps Newly Packaged Genomes in Podoviridae Virions.
J.Biol.Chem., 291, 2016
|
|
4ZKP
 
 | | P22 Tail Needle Gp26 crystallized at pH 7.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tail needle protein gp26 | | Authors: | Sankhala, R.S, Cingolani, G. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Plasticity of the Protein Plug That Traps Newly Packaged Genomes in Podoviridae Virions.
J.Biol.Chem., 291, 2016
|
|
