1IA6
 
 | | CRYSTAL STRUCTURE OF THE CELLULASE CEL9M OF C. CELLULOLYTICUM | | Descriptor: | CALCIUM ION, CELLULASE CEL9M, NICKEL (II) ION, ... | | Authors: | Parsiegla, G, Belaich, A, Belaich, J.P, Haser, R. | | Deposit date: | 2001-03-22 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cellulase Cel9M enlightens structure/function relationships of the variable catalytic modules in glycoside hydrolases.
Biochemistry, 41, 2002
|
|
1K72
 
 | | The X-ray Crystal Structure Of Cel9G Complexed With cellotriose | | Descriptor: | CALCIUM ION, Endoglucanase 9G, GLYCEROL, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1EHX
 
 | | NMR SOLUTION STRUCTURE OF THE LAST UNKNOWN MODULE OF THE CELLULOSOMAL SCAFFOLDIN PROTEIN CIPC OF CLOSTRIDUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDIN PROTEIN | | Authors: | Mosbah, A, Belaich, A, Bornet, O, Belaich, J.P, Henrissat, B, Darbon, H. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum.
J.Mol.Biol., 304, 2000
|
|
1G87
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOCELLULASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-16 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1GA2
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM COMPLEXED WITH CELLOBIOSE | | Descriptor: | ACETIC ACID, CALCIUM ION, ENDOGLUCANASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-29 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1IA7
 
 | | CRYSTAL STRUCTURE OF THE CELLULASE CEL9M OF C. CELLULOLYTICIUM IN COMPLEX WITH CELLOBIOSE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULASE CEL9M, ... | | Authors: | Parsiegla, G, Belaich, A, Belaich, J.P, Haser, R. | | Deposit date: | 2001-03-22 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the cellulase Cel9M enlightens structure/function relationships of the variable catalytic modules in glycoside hydrolases.
Biochemistry, 41, 2002
|
|
1F9D
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellotetraose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FAE
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1F9O
 
 | | Crystal structure of the cellulase Cel48F from C. Cellulolyticum with the thiooligosaccharide inhibitor PIPS-IG3 | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-iodophenyl 1,4-dithio-beta-D-glucopyranoside | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-11 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FBO
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiitol | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1FBW
 
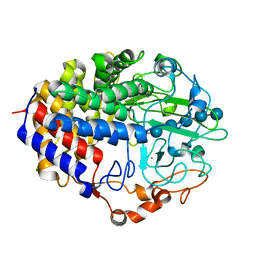 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
1G9G
 
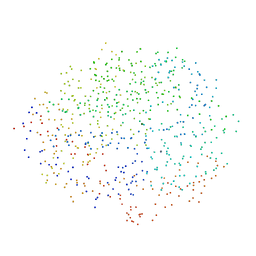 | | XTAL-STRUCTURE OF THE FREE NATIVE CELLULASE CEL48F | | Descriptor: | CALCIUM ION, CELLULASE CEL48F, MAGNESIUM ION | | Authors: | Parsiegla, G, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-11-23 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
1G9J
 
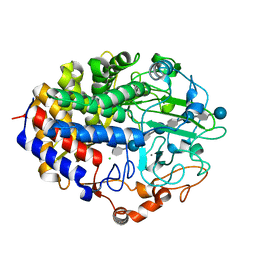 | | X-TAL STRUCTURE OF THE MUTANT E44Q OF THE CELLULASE CEL48F IN COMPLEX WITH A THIOOLIGOSACCHARIDE | | Descriptor: | CALCIUM ION, CELLULASE CEL48F, CHLORIDE ION, ... | | Authors: | Parsiegla, G, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-11-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mutants of cellulase Cel48F of Clostridium cellulolyticum in complex with long hemithiocellooligosaccharides give rise to a new view of the substrate pathway during processive action
J.Mol.Biol., 375, 2008
|
|
1EDG
 
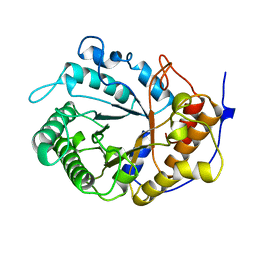 | |
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
1G43
 
 | | CRYSTAL STRUCTURE OF A FAMILY IIIA CBD FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, SCAFFOLDING PROTEIN, ZINC ION | | Authors: | Shimon, L.J.W, Pages, S, Belaich, A, Belaich, J.-P, Bayer, E.A, Lamed, R, Shoham, Y, Frolow, F. | | Deposit date: | 2000-10-26 | | Release date: | 2000-12-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FCE
 
 | | PROCESSIVE ENDOCELLULASE CELF OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, CELLULASE CELF, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Parsiegla, G, Juy, M, Haser, R. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 A resolution.
EMBO J., 17, 1998
|
|
