1GPJ
 
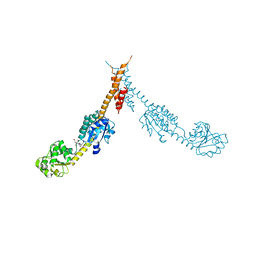 | | Glutamyl-tRNA Reductase from Methanopyrus kandleri | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, CITRIC ACID, GLUTAMIC ACID, ... | | Authors: | Moser, J, Schubert, W.-D, Beier, V, Bringemeier, I, Jahn, D, Heinz, D.W. | | Deposit date: | 2001-11-05 | | Release date: | 2002-01-04 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
EMBO J., 20, 2001
|
|
1O6T
 
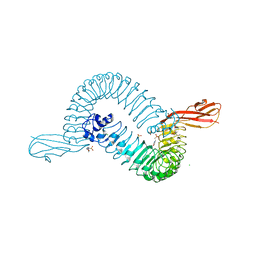 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-15 | | Release date: | 2002-12-23 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
1O6S
 
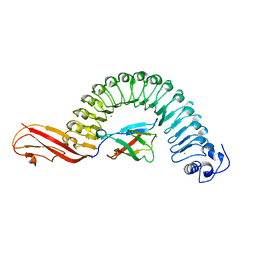 | | Internalin (Listeria monocytogenes) / E-Cadherin (human) Recognition Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, E-CADHERIN, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-13 | | Release date: | 2002-12-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
1O6V
 
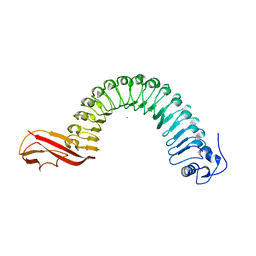 | | Internalin (INLA, Listeria monocytogenes) - functional domain, uncomplexed | | Descriptor: | CALCIUM ION, INTERNALIN A | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-23 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
7QQY
 
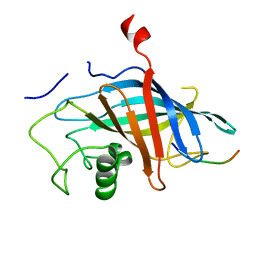 | | yeast Gid10 bound to Art2 Pro/N-degron | | Descriptor: | CHLORIDE ION, ECM21, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A GID E3 ligase assembly ubiquitinates an Rsp5 E3 adaptor and regulates plasma membrane transporters.
Embo Rep., 23, 2022
|
|
8Q7E
 
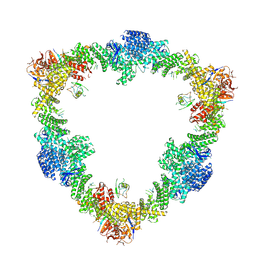 | |
8Q7H
 
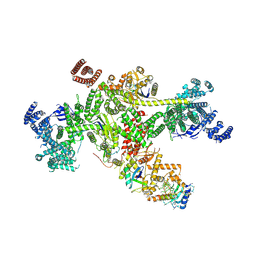 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
8RHZ
 
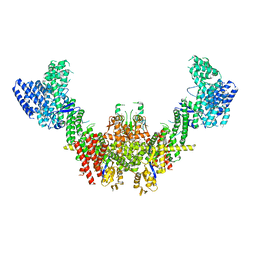 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
6SWY
 
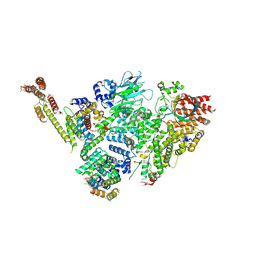 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
8PJN
 
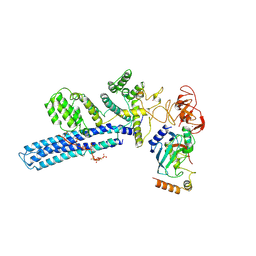 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | Descriptor: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PMQ
 
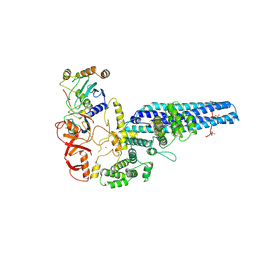 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
