8JYR
 
 | |
8JYQ
 
 | |
3L85
 
 | |
3ACA
 
 | |
3AC9
 
 | |
5XCR
 
 | |
3W6D
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6B
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 | | Descriptor: | GLYCEROL, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6E
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6F
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6C
 
 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3WC3
 
 | | Crystal structure of endo-1,4-beta-glucanase from Eisenia fetida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CITRATE ANION, ... | | Authors: | Arimori, T, Tamada, T. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of endo-1,4-beta-glucanase from Eisenia fetida
J.SYNCHROTRON RADIAT., 20, 2013
|
|
7CEC
 
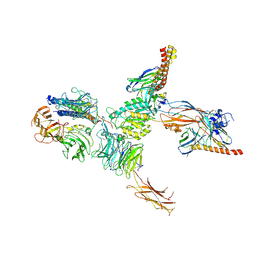 | | Structure of alpha6beta1 integrin in complex with laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arimori, T, Miyazaki, N, Takagi, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of laminin recognition by integrin.
Nat Commun, 12, 2021
|
|
7CEA
 
 | |
7CEB
 
 | |
5XCX
 
 | | Crystal structure of TS2/16 Fv-clasp fragment | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCS
 
 | | Crystal structure of 12CA5 Fv-clasp fragment with its antigen peptide | | Descriptor: | HA peptide, SULFATE ION, VH-SARAH(Y35C) chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCQ
 
 | | Crystal structure of P20.1 Fv-clasp fragment with its antigen peptide | | Descriptor: | C8 peptide, SULFATE ION, VH-SARAH(Y35C)chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.313 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCU
 
 | | Crystal structure of 12CA5 Fv-clasp fragment with its antigen peptide | | Descriptor: | HA peptide, SULFATE ION, VH(S112C)-SARAH chimera,VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCV
 
 | | Crystal structure of NZ-1 Fv-clasp fragment with its antigen peptide | | Descriptor: | CALCIUM ION, PA14 peptide, VH(S112C)-SARAH chimera,VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCT
 
 | | Crystal structure of P20.1 Fv-clasp fragment with its antigen peptide | | Descriptor: | C8 peptide, CHLORIDE ION, VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
7DMU
 
 | |
5GKR
 
 | | Crystal structure of SLE patient-derived anti-DNA antibody in complex with oligonucleotide | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), IgG2, Fab (heavy chain), ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
5GKS
 
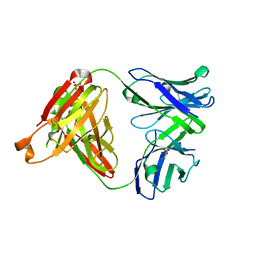 | | Crystal structure of SLE patient-derived anti-DNA antibody | | Descriptor: | IgG2, Fab (heavy chain), PHOSPHATE ION, ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
6LZ4
 
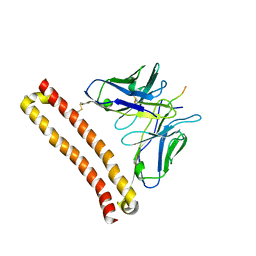 | |
