6GXV
 
 | | Amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, A-amylase, CALCIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GYA
 
 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5FJJ
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
2YEY
 
 | | Crystal structure of the allosteric-defective chaperonin GroEL E434K mutant | | Descriptor: | 60 KDA CHAPERONIN | | Authors: | Cabo-Bilbao, A, Mechaly, A.E, Agirre, J, Spinelli, S, Sot, B, Muga, A, Guerin, D.M.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystal Structure of the Temperature-Sensitive and Allosteric-Defective Chaperonin Groele461K.
J.Struct.Biol., 155, 2006
|
|
5L6S
 
 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
5L6V
 
 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.667 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
5AOG
 
 | | Structure of Sorghum peroxidase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, CALCIUM ION, CATIONIC PEROXIDASE SPC4, ... | | Authors: | Kwon, H, Nnamchi, C.I, Parkin, G, Efimov, I, Agirre, J, Basran, J, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2015-09-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural and Spectroscopic Characterisation of a Heme Peroxidase from Sorghum.
J.Biol.Inorg.Chem., 21, 2016
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
3NAP
 
 | | Structure of Triatoma Virus (TrV) | | Descriptor: | Capsid protein | | Authors: | Squires, G, Pous, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of Triatoma Virus (TrV) highlights the Dicistroviridae and Picornaviridae family differences
To be Published
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-28 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
3ZWG
 
 | | Crystal structure of the pore-forming toxin FraC from Actinia fragacea (form 2) | | Descriptor: | FRAGACEATOXIN C | | Authors: | Mechaly, A.E, Bellomio, A, Morante, K, Gonzalez-Manas, J.M, Guerin, D.M.A. | | Deposit date: | 2011-07-29 | | Release date: | 2012-07-04 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pores of the Toxin Frac Assemble Into 2D Hexagonal Clusters in Both Crystal Structures and Model Membranes.
J.Struct.Biol., 180, 2012
|
|
3ZWJ
 
 | | CRYSTAL STRUCTURE OF THE PORE-FORMING TOXIN FRAC FROM ACTINIA FRAGACEA (Form 3) | | Descriptor: | FRAGACEATOXIN C | | Authors: | Mechaly, A.E, Bellomioa, A, Morantea, K, Gonzalez-Manas, J.M, Guerin, D.M.A. | | Deposit date: | 2011-08-01 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Pores of the Toxin Frac Assemble Into 2D Hexagonal Clusters in Both Crystal Structures and Model Membranes.
J.Struct.Biol., 180, 2012
|
|
6JPH
 
 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | ACETATE ION, Alginate lyase, CALCIUM ION, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
6JPN
 
 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | Alginate lyase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
6JP4
 
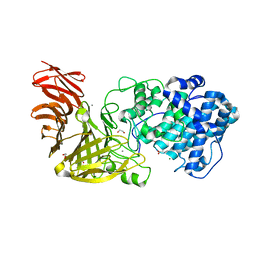 | | Crystal structure of the catalytic domain of a multi-domain alginate lyase Dp0100 from thermophilic bacterium Defluviitalea phaphyphila | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alginate lyase, ... | | Authors: | Ji, S.Q, Dix, S.R, Aziz, A, Sedelnikova, S.E, Li, F.L, Rice, D.W. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | The molecular basis of endolytic activity of a multidomain alginate lyase fromDefluviitalea phaphyphila, a representative of a new lyase family, PL39.
J.Biol.Chem., 294, 2019
|
|
