1AMK
 
 | | LEISHMANIA MEXICANA TRIOSE PHOSPHATE ISOMERASE | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSE PHOSPHATE ISOMERASE | | Authors: | Williams, J.C, Wierenga, R. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power.
Protein Eng., 12, 1999
|
|
2PTK
 
 | | CHICKEN SRC TYROSINE KINASE | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Williams, J.C, Wierenga, R. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-24 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions
J.Mol.Biol., 274, 1997
|
|
2PG1
 
 | |
1WP0
 
 | | Human SCO1 | | Descriptor: | SCO1 protein homolog | | Authors: | Williams, J.C, Sue, C, Banting, G.S, Yang, H, Glerum, D.M, Hendrickson, W.A, Schon, E.A. | | Deposit date: | 2004-08-27 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human SCO1: IMPLICATIONS FOR REDOX SIGNALING BY A MITOCHONDRIAL CYTOCHROME c OXIDASE "ASSEMBLY" PROTEIN
J.Biol.Chem., 280, 2005
|
|
1YGT
 
 | |
5U6A
 
 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-PEG3-MEDITOPE | | Descriptor: | Heavy Chain, Immunoglobulin G binding protein A, Light Chain, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-PEG3-Meditope
To Be Published
|
|
5U5M
 
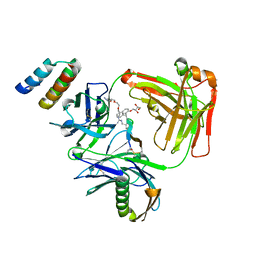 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-MEDITOPE | | Descriptor: | AZIDO-PEG4-MEDITOPE, Immunoglobulin G binding protein A, MEMAB TRASTUZUMAB, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-Meditope
To Be Published
|
|
8G2M
 
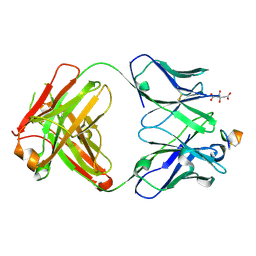 | | The tumor activated anti-CTLA-4 monoclonal antibody XTX101 demonstrates tumor-growth inhibition and tumor-selective pharmacodynamics in mouse models of cancer | | Descriptor: | CITRIC ACID, Heavy chain of humanized IgG, Light chain of humanized IgG, ... | | Authors: | Williams, J.C, Williams, J.C. | | Deposit date: | 2023-02-05 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | XTX101, a tumor-activated, Fc-enhanced anti-CTLA-4 monoclonal antibody, demonstrates tumor-growth inhibition and tumor-selective pharmacodynamics in mouse models of cancer.
J Immunother Cancer, 11, 2023
|
|
8G8N
 
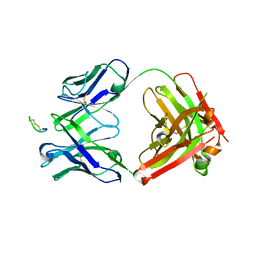 | | CTLA4 Fab with peptide | | Descriptor: | CYS-PRO-GLY-LYS-GLY-LEU-PRO-SER-CYS, Fab heavy chain, Fab light chain | | Authors: | Williams, J.C. | | Deposit date: | 2023-02-18 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | XTX101, a tumor-activated, Fc-enhanced anti-CTLA-4 monoclonal antibody, demonstrates tumor-growth inhibition and tumor-selective pharmacodynamics in mouse models of cancer.
J Immunother Cancer, 11, 2023
|
|
3GC3
 
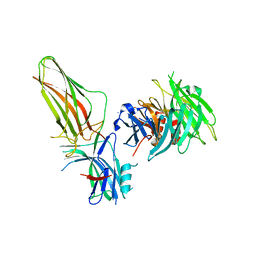 | | Crystal Structure of Arrestin2S and Clathrin | | Descriptor: | Beta-arrestin-1, Clathrin heavy chain 1 | | Authors: | Williams, J.C, Kang, D.S. | | Deposit date: | 2009-02-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking.
J.Biol.Chem., 284, 2009
|
|
1JDL
 
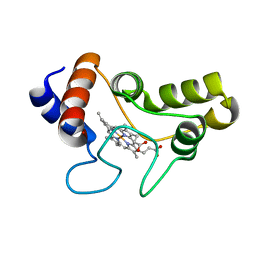 | | Structure of cytochrome c2 from Rhodospirillum Centenum | | Descriptor: | CYTOCHROME C2, ISO-2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Camara-Artigas, A, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-14 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of cytochrome c2 from Rhodospirillum centenum.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2M02
 
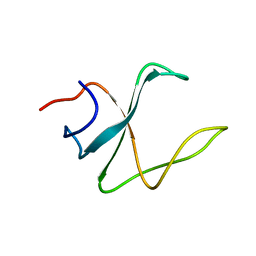 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
5EUK
 
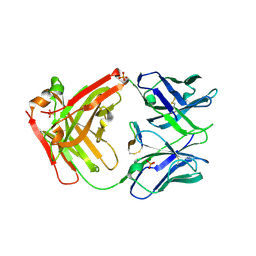 | | Cetuximab Fab in complex with F3H meditope variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ESQ
 
 | | Cetuximab Fab in complex with cyclic beta-alanine-linked meditope | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, Cyclic beta-alanine-linked meditope, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-06-15 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ETU
 
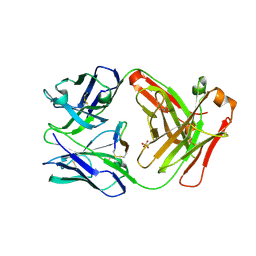 | | Cetuximab Fab in complex with L5E meditope variant | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, L5E meditope variant, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5F88
 
 | | Cetuximab Fab in complex with L5Y meditope variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-12-09 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5FF6
 
 | | Cetuximab Fab in complex with L10Q meditope variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
1JGZ
 
 | | Photosynthetic Reaction Center Mutant With Tyr M 76 Replaced With Lys | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JH0
 
 | | Photosynthetic Reaction Center Mutant With Glu L 205 Replaced to Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGW
 
 | | Photosynthetic Reaction Center Mutant With Thr M 21 Replaced With Leu | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGY
 
 | | Photosynthetic Reaction Center Mutant With Tyr M 76 Replaced With Phe | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGX
 
 | | Photosynthetic Reaction Center Mutant With Thr M 21 Replaced With Asp | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6AU5
 
 | |
6AYN
 
 | |
6AZK
 
 | |
